Researchers in the UK and the United States have developed a stem-cell-based screening platform for identifying compounds that inhibit infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the causative agent for coronavirus disease 2019 (COVID-19).
The platform provides qualitative and quantitative screening using a pseudotyped viral system and stem cell-derived cardiomyocytes expressing the host proteins that SARS-CoV-2 uses for viral entry.
The team – from the University of Cambridge, the University of Colorado, and Addenbrooke’s Biomedical Campus in Cambridge – showed that the platform identified both clinically approved and novel compounds that target these host proteins and inhibit the infection process.
Thomas Williams and colleagues point out that the human embryonic stem cell (hESC)-derived cardiomyocytes used in the platform are clinically relevant and widely available.
Furthermore, the pseudotyped viral system can be used at intermediate biosafety levels meaning that independent research groups could use the platform without requiring higher-level facilities.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
Mortality is higher among SARS-CoV-2 patients with cardiovascular morbidity
The case fatality rate among SAR-CoV-2 patients increases from 2.3% to 10.5% among those with cardiovascular comorbidities. Studies of induced pluripotent stem cell (iPSC)-derived cardiomyocytes have shown that the virus causes direct damage to cardiac tissue.
The SARS-CoV-2 infection process involves the high-affinity binding of a viral surface protein called spike to the host cell receptor angiotensin-converting enzyme 2 (ACE2).
Other host cell proteins are also involved. These include the transmembrane protease serine 2 (TMPRSS2), which mediates spike priming; the protease furin, which proteolytically cleaves spike, and the cathepsins, which carry out endosomal processing. A neutral amino acid transporter called B0AT1 has also been shown to form a complex with ACE2 that can simultaneously bind two spike proteins.
Williams and colleagues previously showed that the genes encoding expression of these proteins are significantly upregulated in the cardiomyocytes of aged individuals, which may explain the increased susceptibility to SARS-CoV-2 among the elderly, suggests the team.
What did the researchers do?
The team set out to determine whether human embryonic stem cell-derived cardiomyocytes (hESC-CMs) also expresses this repertoire of proteins and enable SARS-CoV-2 viral entry
The researchers also investigated whether this model could be used to screen for therapeutic agents that inhibit the infection process.
Using immunohistochemistry to confirm the presence of host cell proteins associated with SARS-CoV-2 infection, the researchers found that more than 95% of the hESC-CMs showed immunolabeling for ACE2, TMPRSS2, furin and cathepsin L, and 27.6% showed immunolabeling for B0AT1.
The researchers then infected the cells with SARS-CoV-2, which resulted in titer- and time-dependent levels of infection.
Creating a drug-screening platform
Next, the team developed a drug-screening platform using the hESC-CMs and a SARS-CoV-2 spike pseudotyped lentivirus expressing green fluorescence protein.
Fluorescent confocal images of 96 well plates showed that the hESC-CMs exhibited an infection rate of 65%.
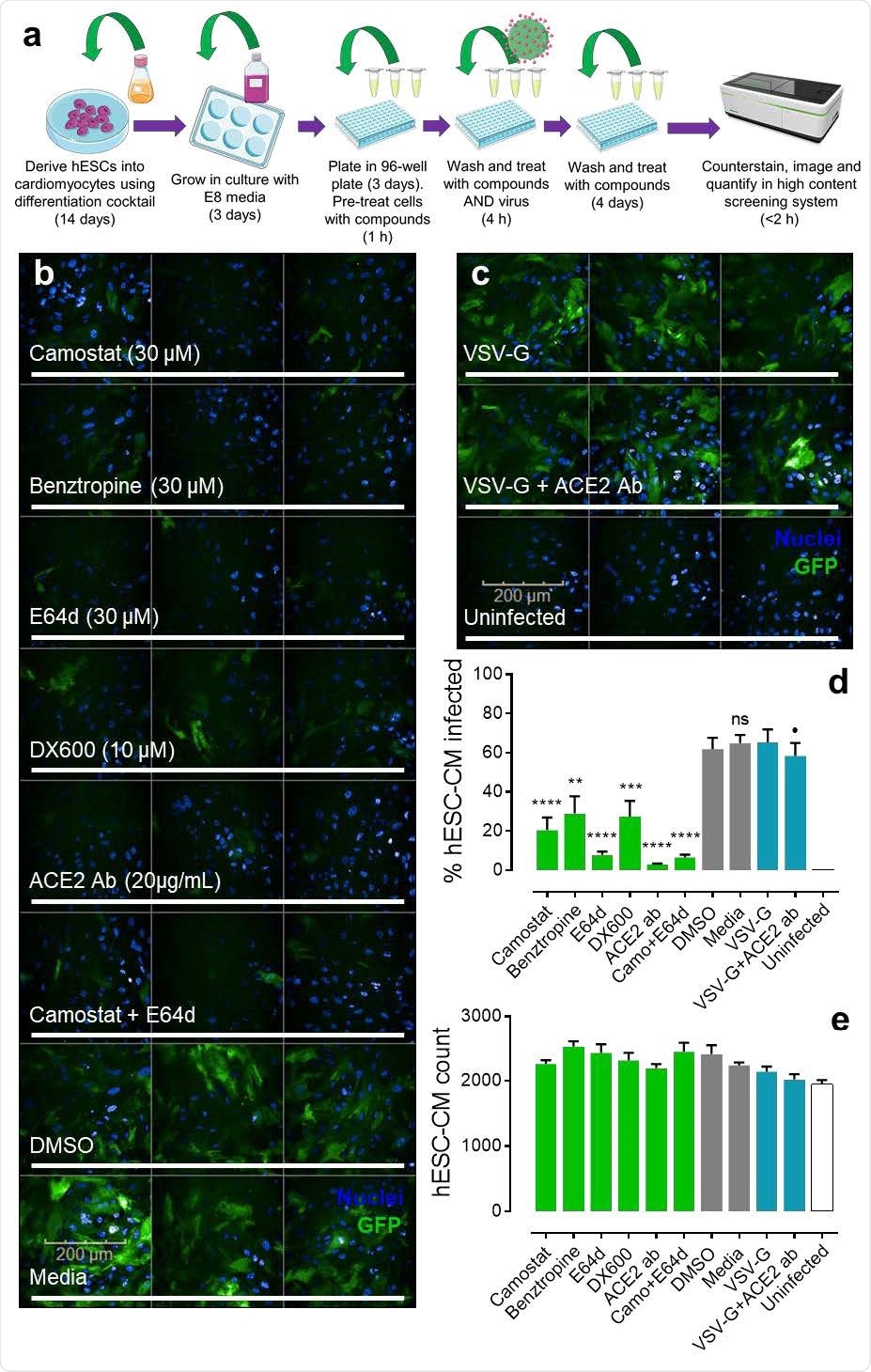
Pseudotyped lentiviral infection in hESC-CMs. a, Schematic showing the experimental workflow in brief for generating human embryonic stem cell-derived cardiomyocytes (hESC-CMs) and taking them into the pseudotyped lentiviral infection drug screen before conducting quantitative imaging (see methods for further details). Schematic was generated using templates from Servier Medical Art. b, Representative fluorescent confocal images (n=2 independent experiments performed in triplicate) of hESC-CMs pre-treated with small molecule inhibitors (camostat, benztropine, E64d), peptide antagonist (DX600), or antibody (ACE2 Ab) targeting protein components involved in SARS-CoV-2 infection. Control cells were treated with DMSO (0.6 %) or media. Cells were treated with drugs for 1 h before incubation with SARS-CoV-2 spike pseudotyped GFP-expressing (green) lentivirus for 4 h. After removal of viral particles, cells were washed and maintained in the presence of drugs for 5 days before fixation with 4 % formaldehyde and staining with Hoechst 33342 nuclear marker (blue). Scale bar shows 200 μm. c, Representative fluorescent confocal images (n=2 independent experiments performed in triplicate) of control human embryonic stem cell-derived cardiomyocytes (hESC-CMs) treated with VSV-G pseudotyped GFP-expressing (green) lentivirus, in the absence (upper) or presence (middle) of antibody (ACE2 Ab). Uninfected controls were not treated with viral particles (bottom). Again, cells were stained with Hoechst 33342 nuclear marker. d, Graphical data showing the percentage of observed hESC-CMs infected with either SARS-CoV-2 spike or VSV-G (control) pseudotyped lentivirus in the presence of drugs or DMSO (0.6 %) as indicated. Uninfected cells were not treated with viral particles. ** = p<0.005; *** = p<0.0005; **** = p<0.00005; and ns = no significant difference (as determined by one-way ANOVA) for each condition versus the DMSO treated control cells. ⚫ = no significant difference for condition versus the VSV-G control. e, Graphical data showing the overall count of observed hESC-CMs for each condition, as indicated. No condition showed a count significantly different (as determined by one-way ANOVA) from the DMSO treated control cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team observed significantly reduced infection levels among wells treated with drugs known to target the host cell proteins involved in SARS-CoV-2 entry and processing.
An ACE2 antibody reduced the infection level to 2.9% and an ACE2 peptide antagonist called DX600 reduced it to 20.5%.
A TMPRSS2 inhibitor called camostat reduced infection to 20.5%, while a cathepsin inhibitor called E64d reduced it to 7.8%.
“These results suggest that inhibition of viral entry can be achieved at distinct steps of viral entry and processing,” says Williams and colleagues.
Importantly, camostat is already clinically approved as a treatment for pancreatitis in Japan and has good safety and tolerability profiles, adds the team.
Interestingly, a small molecule inhibitor of B0AT1, called benztropine, also successfully reduced infection levels (to 28.9%). This drug is already used as an adjunct to parkinsonism therapies, but the precise mechanisms underlying its inhibition of viral infection require further investigation, say the researchers.
A qualitative and quantitative drug-screening platform
“Our results have identified and validated a qualitative and quantitative screen, crucially using hESC-derived beating cardiomyocytes, where these clinically relevant cells are not rate-limiting and widely available,” writes Williams and colleagues.
The team says the findings show that these cells can be used reliably in 96 well plates – the minimum format generally required for high-throughput screening.
“The pseudotyped lentiviral system can also be handled at intermediate Biosafety levels, or containment levels of 2 meaning independent research groups will be able to use this platform without the need for higher-level facilities,” adds the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Williams T, et al. Human embryonic stem cell-derived cardiomyocytes express SARS-CoV-2 host entry proteins: screen to identify inhibitors of infection. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.01.22.427737, https://www.biorxiv.org/content/10.1101/2021.01.22.427737v1
- Peer reviewed and published scientific report.
Williams, Thomas L., Maria T. Colzani, Robyn G. C. Macrae, Emma L. Robinson, Stuart Bloor, Edward J. D. Greenwood, Jun Ru Zhan, et al. 2021. “Human Embryonic Stem Cell-Derived Cardiomyocyte Platform Screens Inhibitors of SARS-CoV-2 Infection.” Communications Biology 4 (1). https://doi.org/10.1038/s42003-021-02453-y. https://www.nature.com/articles/s42003-021-02453-y.