Researchers at the California Institute of Technology in Pasadena have developed a software tool called "Variant Database" that has detected an emerging lineage of severe acute respiratory coronavirus 2 (SARS-CoV-2) isolates in New York.
The novel SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to sweep the globe and has now caused more than 2.41 million deaths.
The Variant Database (VBD) tool can be used to quickly assess the rapidly changing mutational landscape of the viral spike protein that SARS-CoV-2 uses to bind to and infect host cells.
The team says the pattern of mutations in the emerging lineage (now designated B.1.526) suggests that it arose partly in response to selective pressure from antibodies.
"This lineage appeared in late November 2020, and isolates from this lineage account for around 5% of coronavirus genomes sequenced and deposited from New York during late January 2021," writes the team.
Anthony West and colleagues say that based on the collection dates of isolates from this lineage, it appears that they are now increasing in frequency.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
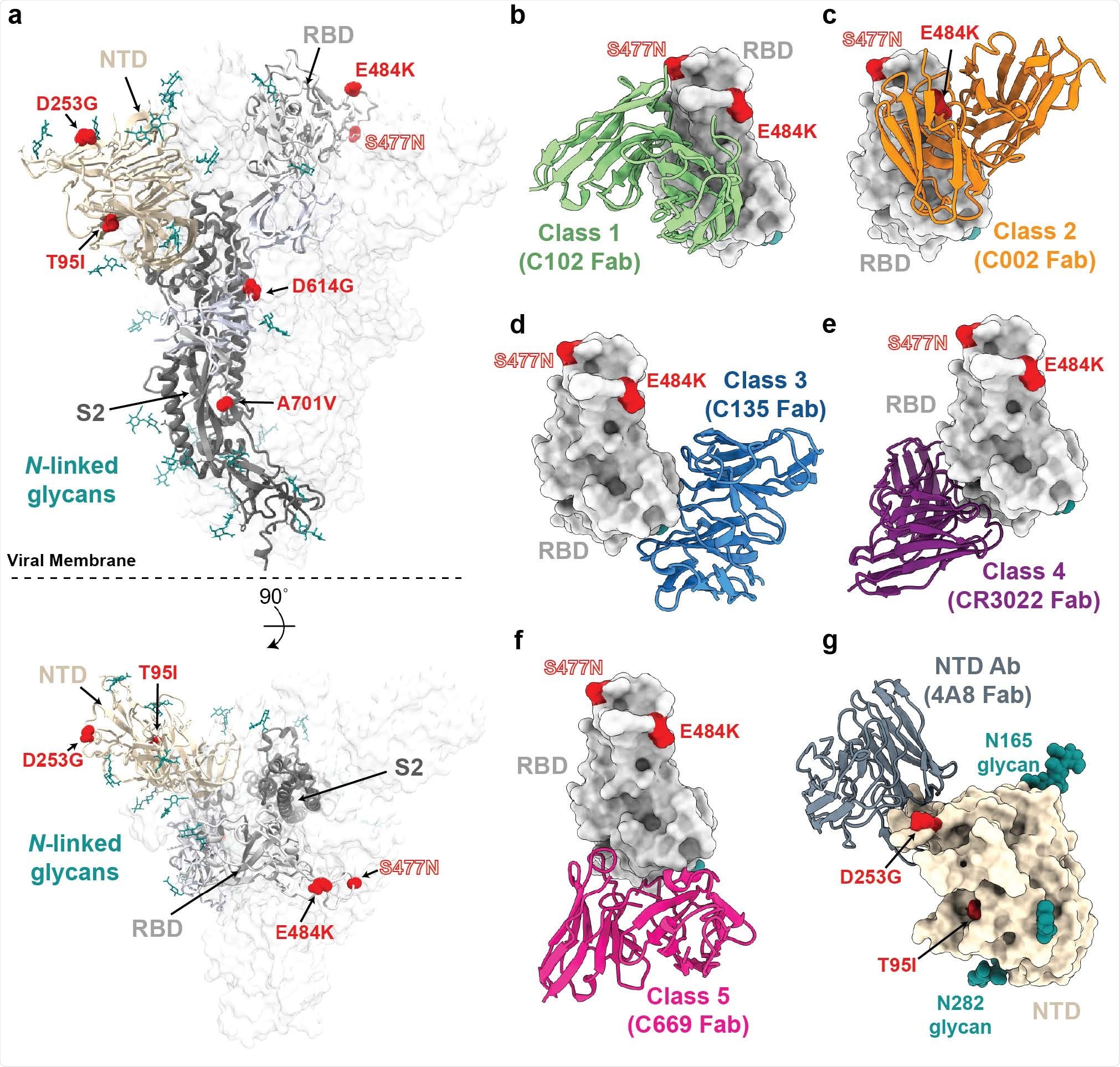
a, Side and top views of the SARS-CoV-2 spike trimer (PDB 7JJI) with mutations of lineage B.1.526 shown as spheres. b-g, Models of representative neutralizing antibodies (cartoon, VH-VL domain only) bound to RBD (b-f, gray surface) or NTD (g, wheat surface). Sites for B.1.526 lineage mutations are shown as red spheres. The S477N site is also shown for the branch containing this mutation instead of the E484K mutation (see Figure 1); b, Class 1 (PDB 7K8M); c, Class 2 (PDB 7K8S); d, Class 3 (PDB 7K8Z); e, Class 4 (PDB 6W41); f, Class 5 (Wang et al., 2021); g, NTD-specific antibody 4A8 (PDB 7C2L).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The variants that have emerged since early stages of the pandemic
Following the early months of the COVID-19 pandemic, the vast majority of SARS-CoV-2 isolates were found to contain the spike mutation D614G.
Several other variants emerged during the fall of 2020, containing numerous mutations that seemed to be focused on the spike protein.
"Multiple lines of evidence support escape from antibody selective pressure as a driving force for the development of these variants," says West and colleagues.
As concerns grow about the effects these mutations could have on the efficacy of antibody therapies and vaccines developed to protect against COVID-19, wide-scale genomic surveillance has been implemented to help researchers monitor and understand the evolution of SARS-CoV-2.
However, "analysis of this wealth of genomic sequences requires a variety of bioinformatics techniques," say the researchers.
The VDB software tool
The VDB software tool created by West and colleagues enables rapid analysis of the changing mutational landscape within the SARS-CoV-2 spike protein.
The tool consists of one program that examines the pattern of spike mutation in a collection of sequenced isolates and another program that generates a list of spike mutations from a multiple sequence alignment. The tool implements a mutation pattern query language that generates a collection of isolates (a "cluster") and a group of spike mutations (a "pattern").
"Generally, clusters can be obtained from searches for patterns, and patterns can be found by examining a given cluster," says West and colleagues.
Clusters can be filtered by variables such as geographical location, collection date, the number of mutations or the presence or absence of a mutation pattern.
What did the authors find?
The team detected an emerging lineage of viral isolates in the New York region that shares mutations with previously reported variants. Several clusters of isolates were detected that had mutations at sites in the spike protein known to be associated with resistance to antibodies against SARS-CoV-2.
The lineage (now designated as B.1.526) comprises two main branches: one including the mutation E484K and another including the mutation S477N, both of which are located within the receptor-binding domain of the spike protein.
What is known so far about these mutations?
Research has previously shown that E484K disrupts the neutralization of numerous anti-SARS-CoV-2 antibodies. The mutation has been identified in variant B.1.351 that recently emerged in South Africa and in the B.1.1.248 variant recently identified in Brazil.
The S477N mutation, which has been identified in several earlier variants, lies near the binding site of multiple antibodies. It has been shown to increase viral infectivity by enhancing the binding of the spike protein to its host cell receptor angiotensin-converting enzyme 2 (ACE2).
"The overall pattern of mutations in this cluster suggests that it arose in part in response to selective pressure from antibodies," writes the researchers.
This B.1.526 variant emerged in late November 2020 and based on the collection dates of isolates from this lineage, it appears that they are now increasing in frequency, concludes West and the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
West AP, et al. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.14.431043, https://www.biorxiv.org/content/10.1101/2021.02.14.431043v1
- Peer reviewed and published scientific report.
West, Anthony P., Joel O. Wertheim, Jade C. Wang, Tetyana I. Vasylyeva, Jennifer L. Havens, Moinuddin A. Chowdhury, Edimarlyn Gonzalez, et al. 2021. “Detection and Characterization of the SARS-CoV-2 Lineage B.1.526 in New York.” Nature Communications 12 (1): 4886. https://doi.org/10.1038/s41467-021-25168-4. https://www.nature.com/articles/s41467-021-25168-4.