Researchers in the United States and Finland have used tissues from people who died from coronavirus disease 2019 (COVID-19) to generate detailed cellular maps of the pathogenetic processes that occur within affected organs during severe disease.
The COVID-19 pandemic – caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – has now infected more than 114 million people and claimed the lives of more than 2.53 million.
Aviv Regev from the Broad Institute of MIT and Harvard in Cambridge, Massachusetts and colleagues say the “COVID-19 cell atlas” they created has provided a rare opportunity to assess the impact of severe disease in the same tissue across multiple individuals, as well as different tissues within the same individual.
“Overall, our COVID-19 cell atlas is a foundational dataset to better understand the biological impact of SARS-CoV-2 infection across the human body and empowers the identification of new therapeutic interventions and prevention strategies,” writes the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Host immune responses are highly variable between individuals
The host immune response to infection varies significantly between individuals, with some experiencing mild or even asymptomatic disease, while others develop severe illness and fatal organ failure.
The majority of deaths are the result of acute lung injury and acute respiratory distress syndrome (ARDS) or complications that lead to multiple organ failure.
Progression to ARDS is thought to involve a combination of increasing viral load, cytopathic effects, the translocation of SARS-CoV-2 to pulmonary tissue, and dysregulated immune responses.
Ineffective viral clearance and tissue damage that occurs during the acute phase of illness can impact the lung, gastrointestinal tract, and various other organs, including the liver, kidney, heart, and brain.
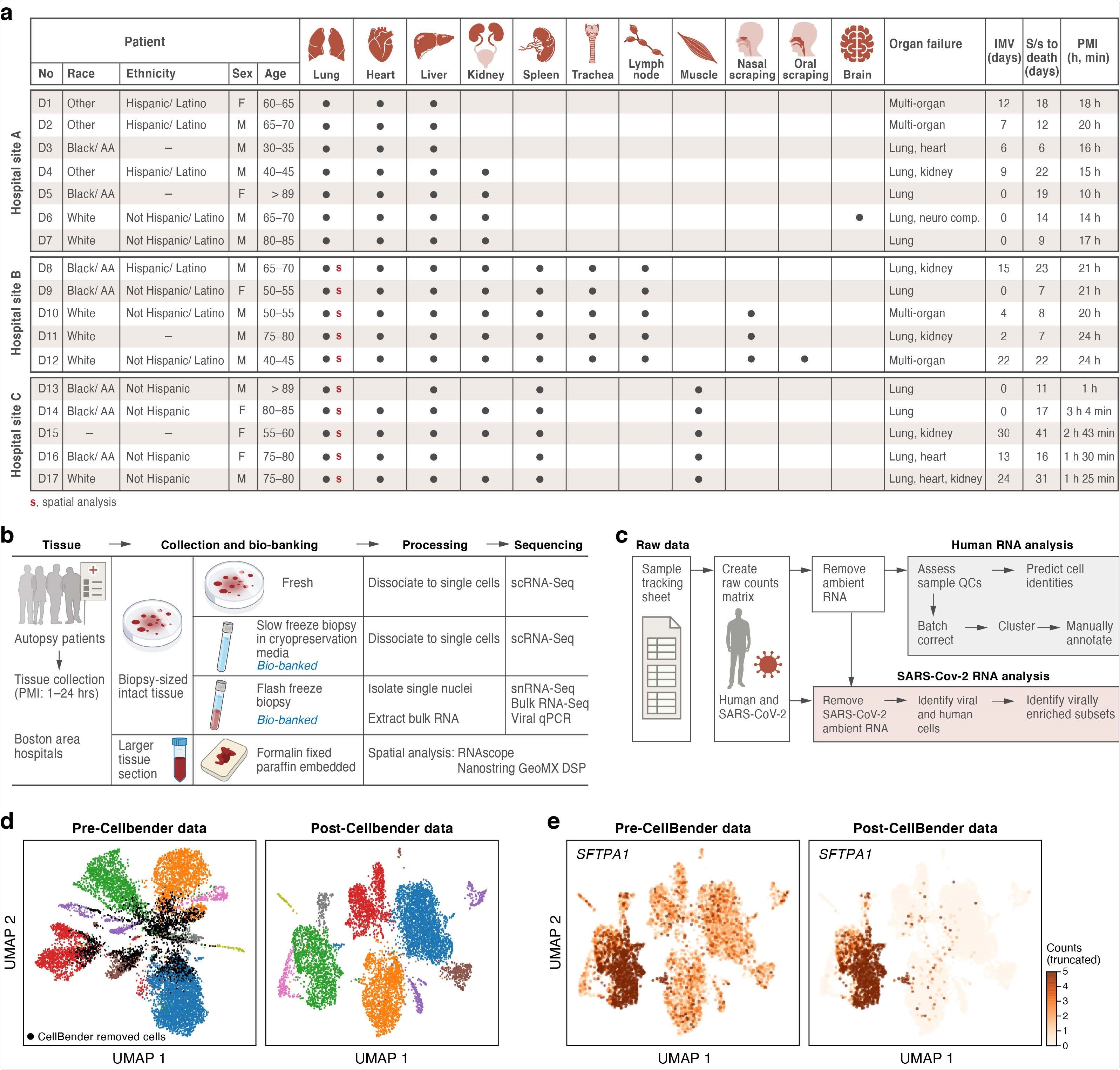
A COVID-19 autopsy cohort for a single cell and spatial atlas a. Cohort overview. IMV: intermittent mandatory ventilation days, S/s: time from symptom onset to death in days; PMI: post-mortem interval. Red bold s: donors for which we collected spatial profiles in the lung. b. Sample processing pipeline overview. c. sc/snRNA-Seq analysis pipeline overview. d,e. CellBender ‘remove-background’ improves cell clustering and expression specificity by removing ambient RNA and empty (non-cell) droplets. UMAP plot of sc/snRNASeq profiles (dots) either before (left) or after (right) CellBender processing, colored by clusters and by doublet status (black) (d), or by expression of the surfactant protein SFTPA1 (e). Color scale in e is linear and truncated at 5 counts to visualize small counts.
Knowledge is still limited in certain areas
Although clinical understanding of severe COVID-19 is rapidly progressing, knowledge about the host immune, molecular and cellular responses associated with pathogenesis and death remains limited.
Evidence suggests that severe COVID-19 is characterized by a so-called “cytokine storm” of pro-inflammatory cytokines and an insufficient antiviral interferon response.
However, researchers are still limited in their understanding of the cell types that are infected by the virus, how the cellular composition is altered in affected tissues and how viral infection alters local cellular responses.
“Addressing these questions using relevant human tissue sources will be essential to inform the identification of new therapeutic targets and prevention strategies,” says Regev and colleagues.
What did the researchers do?
The researchers collected tissues from 11 organs during the autopsies of 17 people who died from COVID-19, providing a tissue bank of approximately 420 specimens.
The team used recently developed methods for single-nucleus RNA sequencing (snRNA-Seq) of frozen specimens and for spatial RNA profiling of formalin-fixed paraffin-embedded tissue to generate comprehensive cellular atlases of lung, kidney, liver and heart tissues that could be compared with atlases of healthy tissues.
Focusing on lung tissue, the researchers charted cell composition and transcriptional programs associated with severe COVID-19.
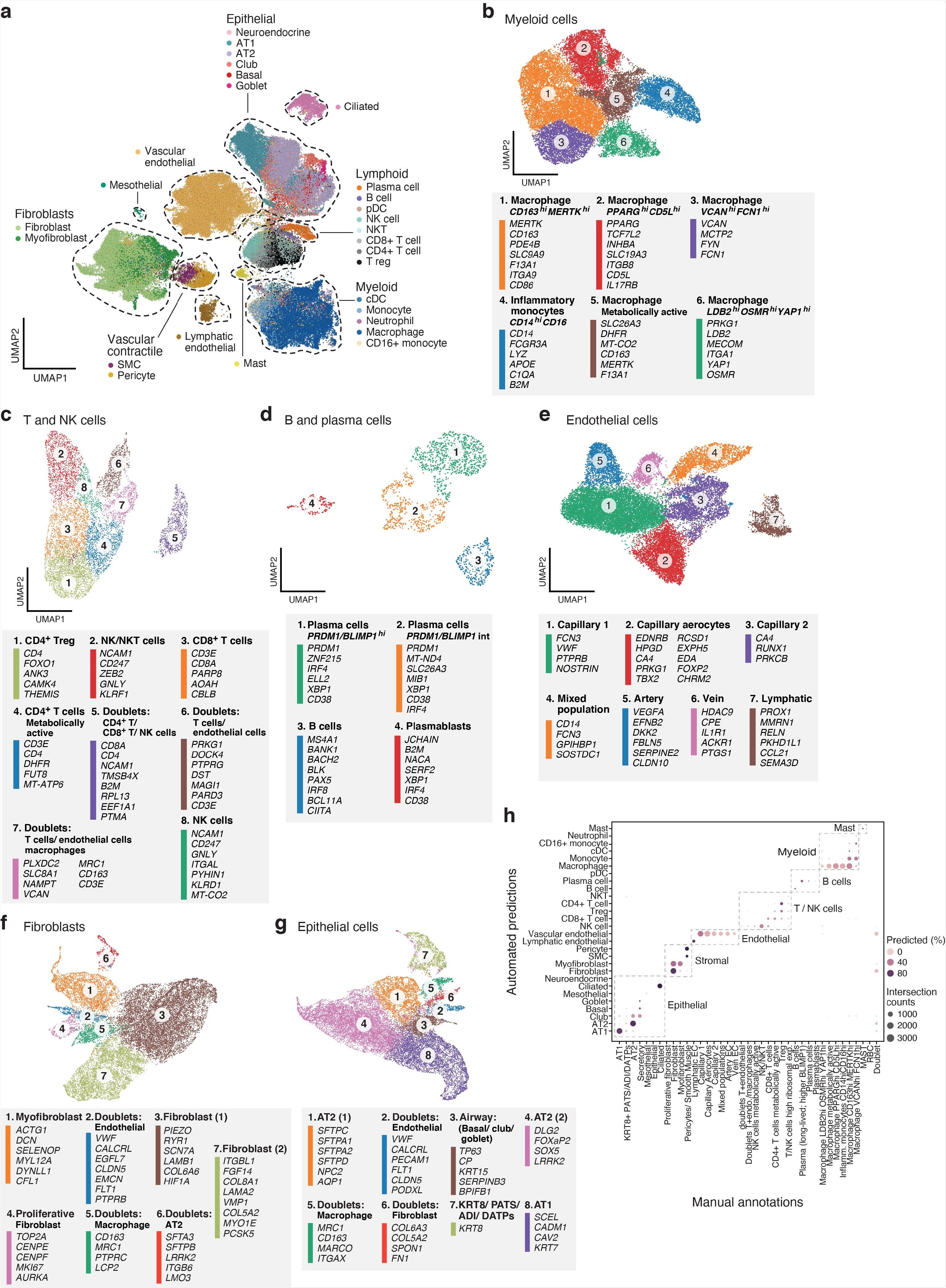
A single cell and single nucleus atlas of COVID-19 lung a. Automatic prediction identifies cells from 28 subsets across epithelial, immune and stromal compartments. UMAP embedding of 106,792 harmonized scRNA-Seq and snRNA-Seq profiles (dots) from all 16 COVID-19 lung donors, colored by their automatically predicted cell type (legend). b-g. Refined annotation of cell subsets within lineages. UMAP embeddings of each selected cell lineages with cells colored by manually annotated sub-clusters. Color legends highlight highly expressed marker genes for select subsets. b. myeloid cells (24,417 cells/ nuclei), c. T and NK cells (9,950), d. B and plasma cells (1,693), e. endothelial cells (20,366), f. fibroblast (20,925), g. epithelial cells (21,700). h. High consistency between automatic and manual annotations. The proportion (color intensity) and number (dot size) of cells with a given predicted annotation (rows) in each manual annotation category (columns)
What did they find?
Compared with healthy lung tissue, the team observed significantly altered transcriptional programs within the epithelial, immune, and stromal compartments of COVID-19 lung tissue, as well as intrinsic changes in multiple cell types.
Analysis of epithelial cell subsets and programs showed that multiple avenues of tissue repair had failed within the diseased lungs.
The team observed evidence of alveolar type 2 (AT2) differentiation replacing depleted AT1 lung epithelial cells, as has previously been observed in fibrosis.
This was accompanied by an increase in myofibroblasts, thereby reflecting defective tissue repair.
The researchers also observed the pre-alveolar type-1 transitional cell state (PATS) and the presence of TP63+ intrapulmonary basal-like progenitor (IPBLP)-like cells, which are similar to the cells seen following infection with H1N1 influenza.
Regev and colleagues say the presence of both PATS and IPBLP-like cells suggests multiple regenerative strategies had been induced to re-establish the alveolar epithelial cells that had been lost to viral infection.
“We hypothesize that the loss of AT2 cells and their insufficient self-replacement may trigger the mobilization of IPBLP as a last-resort strategy to regenerate the alveolar epithelial barrier, though this may come at the cost of fully-functional alveolar structure and function,” they write.
Levels of viable epithelial cells were lower in samples with higher viral load
Compared with samples with lower viral loads, levels of viable epithelial cells, including AT1 and AT2, were significantly lower in seven samples with the highest viral load.
In samples from four individuals with the highest viral load, epithelial cells accounted for less than 10% of the total cells captured.
“This may be due to excessive epithelial death secondary to high viral replication within targeted pneumocytes and a highly inflammatory environment,” write the researchers.
Regev and colleagues say spatial analysis of the COVID-19 lung tissue supported previous reports that high viral levels mainly occur during the earlier stages of infection. The team observed upregulation of early genes such as JUN and FOS only in epithelial regions of the lung with high levels of SARS-CoV-2.
The researchers say the data presented here can be used to inform future studies of COVID-19 tissue pathology and pathophysiology.
“Overall, our atlas provides critical insights into the pathology, pathogenesis and pathophysiology of severe COVID-19 and should help inform future therapeutic development and prophylactics,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Regev A, et al. A single-cell and spatial atlas of autopsy tissues reveals pathology and cellular targets of SARS-CoV-2. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.25.430130, https://www.biorxiv.org/content/10.1101/2021.02.25.430130v1
- Peer reviewed and published scientific report.
Delorey, Toni M., Carly G. K. Ziegler, Graham Heimberg, Rachelly Normand, Yiming Yang, Åsa Segerstolpe, Domenic Abbondanza, et al. 2021. “COVID-19 Tissue Atlases Reveal SARS-CoV-2 Pathology and Cellular Targets.” Nature 595 (7865): 107–13. https://doi.org/10.1038/s41586-021-03570-8. https://www.nature.com/articles/s41586-021-03570-8.