The delivered gene-editing proteins were functional, thereby establishing that VLPs are promising vehicles for therapeutic macromolecule delivery. Furthermore, in a remarkable demonstration of improved visual function in a mouse model of genetic blindness, the scientists also reported undetected ‘off-target effects’ of this treatment approach.

Study: Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Image Credit: metamorworks / Shutterstock.com
Introduction
One way to treat a genetic disease is by directly targeting abnormal single nucleotides through the use of gene editing agents. The most common base editors (BEs) used for this purpose include the cytosine BE and the adenosine BE, both of which can correct known disease-causing single-nucleotide variants.
It is important to target and edit variants without inducing double-stranded deoxyribonucleic acid (DNA) breaks (DSBs), as this simultaneously minimizes indels, large deletions, translocations, chromothripsis, and other chromosomal abnormalities. To date, several studies have successfully used BEs to correct pathogenic point mutations, thus rescuing disease phenotypes in mice and nonhuman primates.
Despite the utility of BEs, several challenges limit the ability to achieve desired targets in vivo with minimal off-target editing. One such challenge includes targeted delivery of these BEs, which has led researchers to investigate several delivery approaches for BEs in vivo, including viruses like adeno-associated viruses (AAVs).
However, using viruses to transport DNA-encoded editing agents is also associated with certain limitations. For example, these delivery vehicles can induce prolonged expression in transduced cells that make the virus delivery have a higher frequency of off-target editing. In addition, there is also an increased possibility of insertional mutagenesis, which is defined as viral vector integration into the genome of transduced cells. Unfortunately, both of these potential adverse effects of viral delivery vehicles can lead to unwanted oncogenesis.
Alternatives to viral delivery approaches
As a result of the limitations of viral delivery, alternative strategies are being explored to deliver gene-editing agents. For example, previous reports have demonstrated successful base editing in the mouse inner ear and retina following local administration of lipid-encapsulated BE proteins or ribonucleoproteins (RNPs).
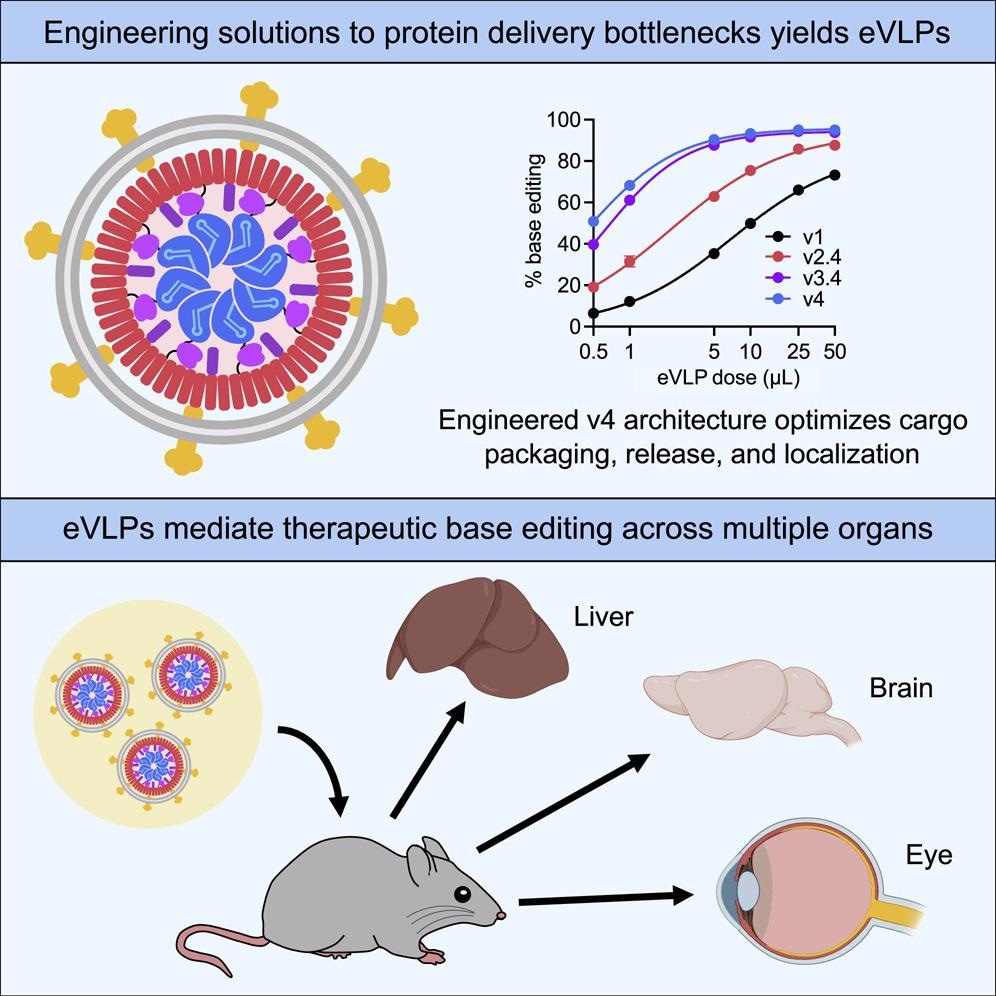
In an effort to further enhance the delivery of BE RNPs to multiple tissues and organs in vivo, the scientists in the present study developed assemblies of inactivated viral proteins that can reach target cells to deliver gene-editing agents. These assemblies, referred to as virus-like particles (VLPs), have emerged as potentially promising vehicles for efficient RNP cargoes that eliminate the risks associated with viral genome integration and prolonged expression of the editing agent.
It should be noted that existing VLP-mediated strategies for delivering gene-editing agent RNPs have only modest editing efficiencies with limited therapeutic efficacy in vivo. Thus, the therapeutic levels of postnatal in vivo gene-editing using RNP-packaging VLPs have not been previously reported.
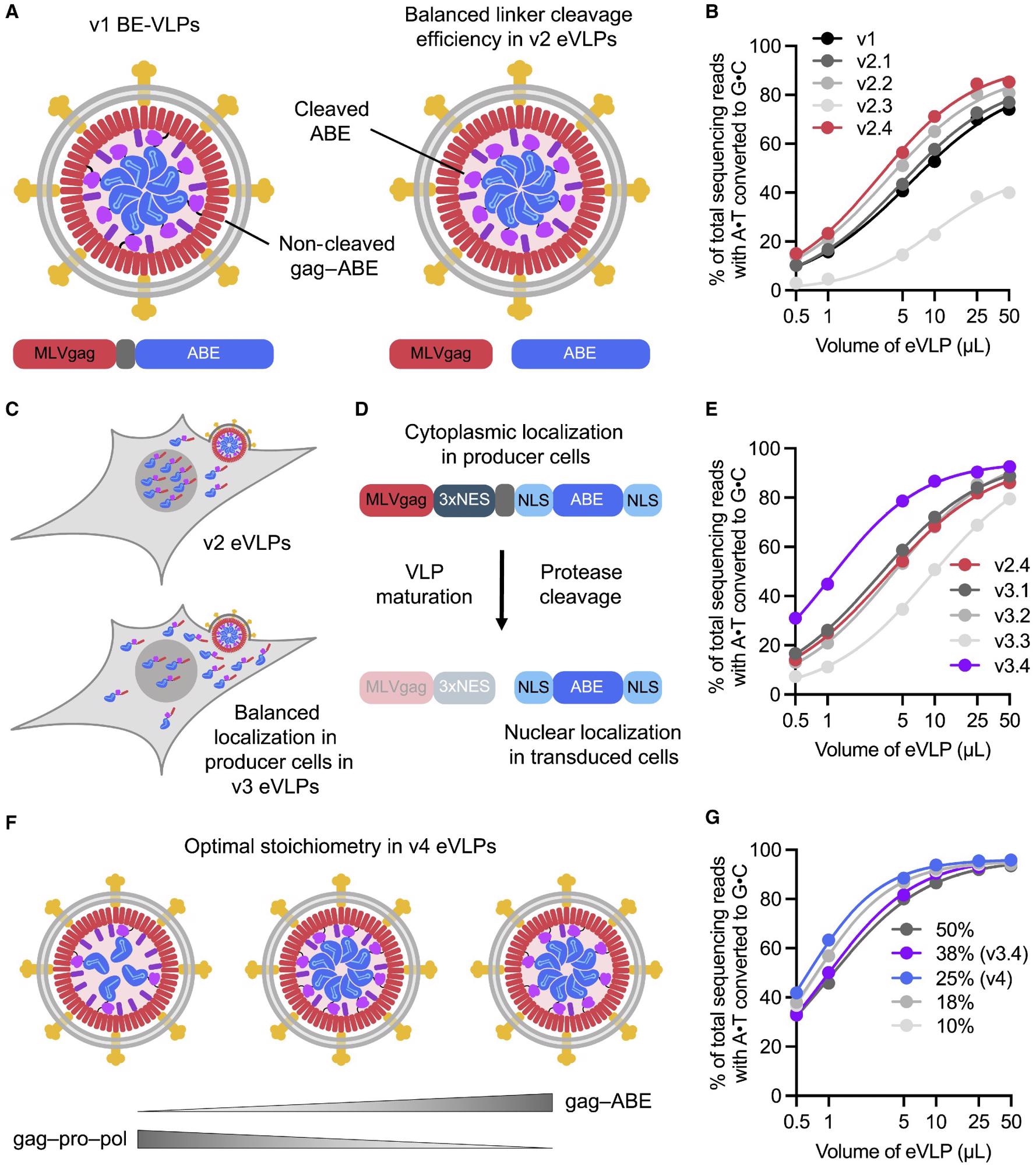
Identifying and engineering solutions to bottlenecks that limit VLP potency results in v2, v3, and v4 eVLPs (A) More efficient linker cleavage leads to improved cargo release after VLP maturation. (B) Adenine base editing efficiencies of v1 and v2 BE-eVLPs at position A7 of the BCL11A enhancer site in HEK293T cells. (C) Improved localization of cargo in producer cells leads to more efficient incorporation into eVLPs. (D) Installing a 3xNES motif upstream of the cleavable linker encourages cytoplasmic localization of gag–3xNES–cargo in producer cells but nuclear localization of free ABE cargo in transduced cells. (E) Adenine base editing efficiencies of v2.4 and v3 BE-eVLPs at position A7 of the BCL11A enhancer site in HEK293T cells. (F) The optimal gag–cargo:gag–pro–pol stoichiometry balances the amount of cargo protein per particle with the amount of MMLV protease required for efficient particle maturation. (G) Adenine base editing efficiencies of v3.4 eVLPs with different gag–ABE:gag–pro–pol stoichiometries at position A7 of the BCL11A enhancer site in HEK293T cells. Legend denotes % gag–ABE plasmid of the total amount of gag–ABE and gag–pro-pol plasmids. (B, E, and G) Values and error bars reflect mean ± SEM of n = 3 biological replicates. Data were fitted to four-parameter logistic curves using nonlinear regression.
In vitro assessment
In the present study, the scientists developed an engineered VLP platform to package and deliver both in vitro and in vivo therapeutic RNPs, including base editors and Cas9 nuclease.
To this end, the researchers designed an extensive VLP that has yielded fourth-generation (v4) eVLPs. This eVLP can carry an average of 16-fold more BE RNPs as compared with previously reported designs. RNPs were delivered in these v4 eVLPs for therapeutically relevant ex vivo and in vivo applications.
The v4 eVLPs developed in the current study were found to be highly efficient in targeted base editing with minimal off-target editing in a variety of cell types. The authors used multiple immortalized cell lines, primary human and mouse fibroblasts, and primary human T-cells to test this delivery efficiency.
Compared to prior generations of eVLPs, the v4 BE-eVLPs were found to exhibit a 5.6 fold improvement of their editing efficiency in vitro. Furthermore, when tested in HEK293T cells, v4 BE-eVLPs provided an editing activity that was 8.5-fold better than prior generations of eVLPs. When tested, similar trends were observed in the enhanced editing efficiency of v4 BE-eVLPs. Various genes were targeted in several other immortalized cell lines and primary human and mouse fibroblasts and primary human T-cells.
In addition to improved editing efficiency, the researchers also evaluated the Cas-dependent off-target editing by v4 BE-eVLPs as compared to plasmid transfection. To this end, comparable or higher on-target editing efficiency was observed in these experiments, thereby confirming the specificity of these particles to only deliver BEs to target genes.
Testing the efficacy of eVLPs in mice
The first series of in vivo tests on the delivery efficiency of these eVLPs involved the injection of v4 BE-eVLPS that would install a silent mutation in mouse Dnmt1, a genomic locus that is amenable to nuclease-mediated indel formation and adenine base editing. These specific eVLPs were co-injected into the cerebrospinal fluid of neonatal mice, which successfully edited 53% and 55% of the cortex and mid-brain cells in these mice, respectively.
When tested for their utility as a therapeutic tool in adult animals, the researchers utilized the v4 BE-eVLPs to target the proprotein convertase subtilisn/kexin type 9 (Pcsk9), which is a therapeutically relevant gene that is involved in the homeostasis of cholesterol levels. Since this strategy was meant to focus on the ability of the eVLPs to specifically knockdown Pcsk9 in the liver, the researchers were also interested in confirming the specificity of their treatment to the liver alone.
To this end, 63% editing in the bulk liver following the highest dose of v4 BE-eVLPs was observed, which confirmed the successful delivery of this gene-editing tool to the liver. Furthermore, minimal editing above background levels was detected in the lungs, kidneys, heart, and muscle; however, 4.3% base editing was observed in the spleen. Taken together, these findings demonstrate that v4 BE-eVLPs offer on-target editing with minimal off-target editing in vivo.
The researchers also sought to determine whether the v4 BE-eVLPs could successfully correct a disease-causing point mutation in an adult mouse model. To this end, the researchers utilized a mouse model that resembles Leber congenital amaurosis (LCA), a family of retinal disorders associated with degeneration of the retinas, early visual impairment, and blindness.
The rd12 mouse model used in this study has a nonsense mutation in exon 3 of Rpe65, which causes almost complete blindness in these mice. Therefore, for the purposes of this experiment, the researchers designed their v4 BE-eVLPs to target the Rpe65(R44X) mutation, which were subsequently injected subretinally into the mice. This experiment demonstrated that at five weeks post-injection, the eVLPs could successfully target the retina and partially restore visual function.
Conclusions
These findings presented in the current study establish eVLPs as a platform for transiently delivering gene editing agents in vivo. Furthermore, the experiments performed here provide evidence for the high therapeutic efficiency and minimal risk of off-target editing or DNA integration associated with this treatment approach.
Journal reference:
- Banskota, S., Raguram, A., Suh, S., et al. (2022) Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell Press. doi:10.1016/j.cell.2021.12.021.