Since the discovery of penicillin in 1928, antibiotics have revolutionized the treatment of infections.
In the 20th century, previously common and deadly diseases, such as tuberculosis, suddenly became curable and, since then, antibiotics have become a mainstay of modern healthcare.
But there are fears that the era in which we can readily treat bacterial infections is drawing to a close due to the rise of antibiotic resistance.
The lowest estimates suggest 700,000 people per year die because of antimicrobial resistance, and this could rise to 10 million per year by 2050.
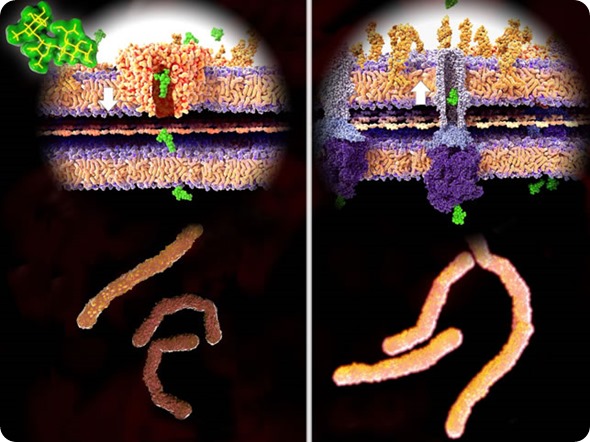
Antibiotic resistance, 3d rendering, left: streptomycin (green) passes the bacterial wall, killing the bacteria. Right: Bacteria have developed resistance and expel strept. through TolC efflux-pump.
© Juan Gaertner / Shutterstock.com
Microbes are increasingly becoming resistant to older medicines and, at the same time, there is an insufficient number of new medicines in the pipeline to overcome resistant organisms.
Improving the way that we use antibiotics to put the brakes on emerging resistance is therefore an essential strategy for easing the pressure to develop new antimicrobials and cutting the risks resistance poses to global health.
Rapid diagnostics at the bedside
Strategies to optimize the use of antibiotics – referred to under the umbrella of ‘antimicrobial stewardship’ – center on identifying the most appropriate antibiotic for the infection, as well as the ideal dose and duration.
But the ability to do this is hampered by the limited availability of rapid diagnostics that can guide clinicians towards the right antibiotic at the right time.
The Independent Review on Antimicrobial Resistance, which was commissioned by the UK Prime Minister to recommend a series of actions to tackle global antimicrobial resistance, identified rapid diagnostic tests as key to enacting behavior change in antibiotic prescriptions.
The review panel call for these “game-changing” diagnostics to undergo widespread adoption between now and 2020 to bring about a “right patient, right antibiotic, right time” approach to antibiotic prescribing.
“If the world is serious about tackling the threat of drug-resistant infections, we need to fully embrace the step-change in technology that rapid point-of-care diagnostics represent,” the review concluded.
“Only by doing this can we fundamentally and sustainably reduce our misuse and overuse of antibiotics.”
Accelerating diagnosis of bloodstream infections
Scientists at the UK-based company, Momentum Bioscience, have created a new method for confirming the absence of bacteraemia and fungaemia infections called the Enzymatic Template Generation and Amplification (ETGA).
ETGA has the potential to enable rapid confirmation of the absence of pathogens in a blood culture with an NPV of 99.5%. The test works by using the fact that DNA polymerase, a major enzyme involved in DNA replication, is produced by all viable microbes.
First, a sample of blood culture suspected to contain fungi or bacteria is processed to separate potential pathogens, which are subsequently lysed so that their enzymes are released.
These are then incubated with the ETGA substrate, a pair of short oligonucleotides. The oligonucleotides, in the presence of the microbe's DNA polymerase, become extended as a result of the enzyme’s native action. In the absence of DNA polymerase, there is no substrate modification. This can be easily measured using real-time polymerase chain reaction, indicating the absence of pathogens with a high NPV.
This method has a number of potential applications. One such use is in the detection of bacteremia in blood samples.
Currently, blood culture is the gold standard for confirming bloodstream infection but one major drawback is the time it takes to detect pathogens: it is currently standard to report a sample negative after five days.
In the interim, patients are usually treated with broad-spectrum antibiotics that are only withdrawn once a negative result comes through – as is the case for 90% of samples.
This means that these patients are being exposed to antibiotic overuse, which not only contributes to antibiotic resistance, but can also cause direct harm to them through antibiotic toxicity and an increased risk of C. difficile infection.
In a 2013 study, Zweitzig et al. showed that the ETGA method is able to detect bacteria from spiked blood culture samples.
They found that, in comparison to the lengthy time taken to detect pathogens using a traditional blood culture method, the ETGA assay detected S. aureus and E. coli after only two hours of incubation.
It was also able to detect very low concentrations of bacteria – at as little as 10 colony-forming units per mL. As the method has a very high negative predictive value, the promising results suggest that ETGA could be used in a clinical setting to inform antibiotic discontinuation within a matter of hours from sample collection.
Dr Helen Bennett reviews the Vitl Ther-Mix
Streamlining methods with the Ther-Mix
The ETGA method is much faster than traditional blood culture, but it can be made even quicker via automation. The team at Momentum use Vitl Life Science Solutions’ Ther-Mix at the center of the ETGA technique to streamline their methods.
By allowing them to semi-automate their workflow, the Ther-Mix helps to deliver results even faster, cut down on hands-on labor, free up bench space and enhance the standardization of their methods.

With a Ther-Mix, multiple mixing and incubation steps can be carried out by one unit with the capability for up to 100 customized mixing programs to be plugged in from start to finish, allowing operators to walk away and let the Ther-Mix do the work.
References
- Crow MA, Bennett HV, Grover M, et al. Evaluation of a new method for the rapid exclusion of infection in suspected bacteraem. Presented at: 24th European Congress of Clinical Microbiology and Infectious Diseases; 10-13 May 2014. Barcelona, Spain.
- Dryden M. Near-patient testing, infection biomarkers, and rapid diagnostics. In: Laundy et al. (eds.) Antimicrobial Stewardship. Oxford University Press, 2016.
- Kopnitsky M (2013). A new twist on the detection of disease-causing microbes. Available at: http://www.mlo-online.com/a-new-twist-on-the-detection-of-disease-causing-microbes.php
- O’Neill J, chair (2014). Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Available at: http://amr-review.org/Publications
- O’Neill J, chair (2015). Rapid diagnostics: Stopping unnecessary use of antibiotics. Available at: http://amr-review.org/Publications
- Zweitzig DR, Sodowich BI, Riccardello NM, et al. Feasibility of a Novel Approach for Rapid Detection of Simulated Bloodstream Infections via Enzymatic Template Generation and Amplification (ETGA)eMediated Measurement of Microbial DNA Polymerase Activity. Journal of Molecular Diagnostics 2013; 15: 319-330.
About Vitl Life Science Solutions

Vitl designs, manufactures and sells high quality laboratory instruments to compete with market leaders at an affordable price with unrivalled functionality, usability and style.
The growing Vitl laboratory products range includes the world’s best-selling microplate heat sealer- the VTS and its compact counterpart the MicroTS. Vitl also sell a range of foil and film heat seals as well as microtube pickers and programmable mixers and vortexers.
Vitl products was established in 2006 by the medical, diagnostic and analytical device design and manufacturing company Integrated Technologies Ltd.
As a member of the ITL group, Vitl benefits from the experience and expertise of a parent company that works with globally successful medical devices and laboratory instrumentation.
Vitl products are sold through a global network of distributors and OEM (private label) partners, managed by their main sales offices in the UK, USA and China.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.