Pseudoviruses are synthetic viruses used to inject genetic material, including DNA and RNA, with specific and desired traits into bacterial and eukaryotic cells. Pseudoviruses are closely related to viruses in structure and behavior but lack many characteristics exhibited by real viruses, including the capability to replicate.
Dianfan Li and colleagues say their strategy for selecting and designing the synthetic nanobodies (sybodies) could be useful for developing therapies to combat coronavirus disease 2019 (COVID-19) and similar outbreaks, should they occur in the future.
“Our work presents an alternative approach to generate neutralizers against newly emerged viruses,” writes the team.
A pre-print version of the paper can be accessed in the server bioRxiv*, while the article undergoes peer review.
Researchers are trying to understand infectivity mechanisms
Since the COVID-19 outbreak first emerged in Wuhan, China, late last year, the pandemic has now infected more than 7.48 million people and killed almost 420,000 worldwide.
Rapid transmission of COVID-19’s causative agent – SARS-CoV-2 – has prompted urgent efforts to shed light on the mechanisms underlying its high infectivity.
Researchers already know that this primarily involves a viral surface structure called the Spike protein, which is cleaved by host cell proteases into two subunits: S1 and S2.
The S1 subunit binds to the host cell surface receptor – angiotensin-converting enzyme 2 (ACE2) – via its receptor-binding domain (RBD) and causes a conformational change that enables S2-mediated fusion at the plasma membrane.
This key role of the RBD in viral attachment has made it a popular target for neutralization approaches that will block its ability to bind ACE2.
Nanobodies and sybodies
Single-domain antibodies or nanobodies derived from the llama have been shown to bind antigens with affinities comparable to those of conventional antibodies, while generally being more heat stable, easier to generate (can be made in bacteria) and easier to genetically engineer than conventional antibodies.
Recently, three libraries of synthetic nanobodies (sybodies) have been designed, from which nano-molar affinity antibodies can quickly be selected and obtained, within as little as two weeks.
“This accelerated pace is most attractive in cases to develop antibodies against newly emerged and quick spreading diseases like Covid-19,” say Li and colleagues. Furthermore, unlike isolating antibodies form convalescent patients, the selection from libraries does not require high-level biosafety procedures, making it more accessible to the broader scientific community.
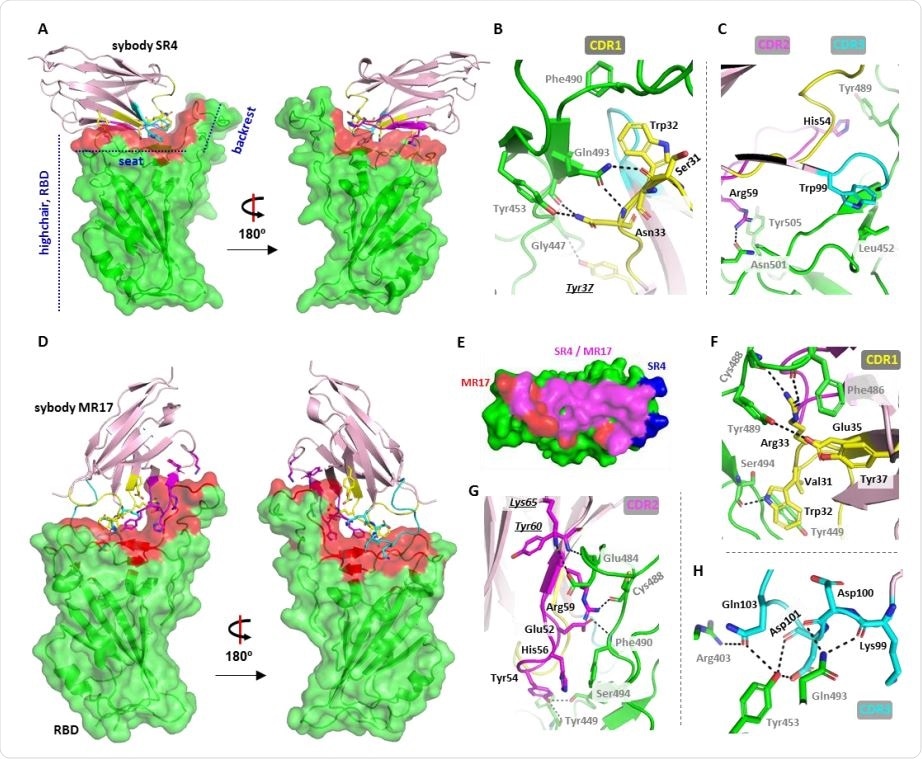
Crystal structure of two sybody-RBD complexes. (A) The overall structure of SR4 (pink cartoon) bound with RBD (green surface) which resembles a short backrest high chair. The binding surface is highlighted red. (B,C) Interactions between SR4 and the RBD (green) contributed by CDR1 (yellow), CDR2 (magenta), and CDR3 (cyan). The framework residue Tyr37 involved in the interactions is shown underlining italic. (D) The overall structure of the MR17-RBD complex. Color-coding is the same as in A. (E) The overlap between the SR4- and MR17-interacting surfaces. (F-H). Interactions between MR17 and the RBD (green) contributed by CDR1 (yellow), CDR2 (magenta), and CDR3 (cyan). The framework Lys65 and Tyr60 participated in the binding are shown underling italic. Dashed lines indicate H-bonding or salt-bridges between atoms that are <4.0 Å apart. Labels for sybody residues are colored black and labels for the RBD residues are colored grey.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Generating unique neutralizing sybodies
Now, Li and the team have produced unique sybodies for neutralizing the Spike (S) protein RBD by selecting SARS-CoV-2 S-RBD binders from three high-diversity sybody libraries.
Eight “first-comers” first selected using an ELISA test were further screened using a fluorescence-detector size exclusion chromatography assay to assess their ability to bind RBD at low concentrations.
This generated nine sybodies of interest for crystallization studies, which further narrowed the selection down to four candidates for biochemical characterization, namely SR4, MR3, MR4, and MR17.
Bio-layer interferometry was then used to test the ability of the sybodies to bind the SARS-CoV-2 RBD, which showed that MR3 had the highest affinity.
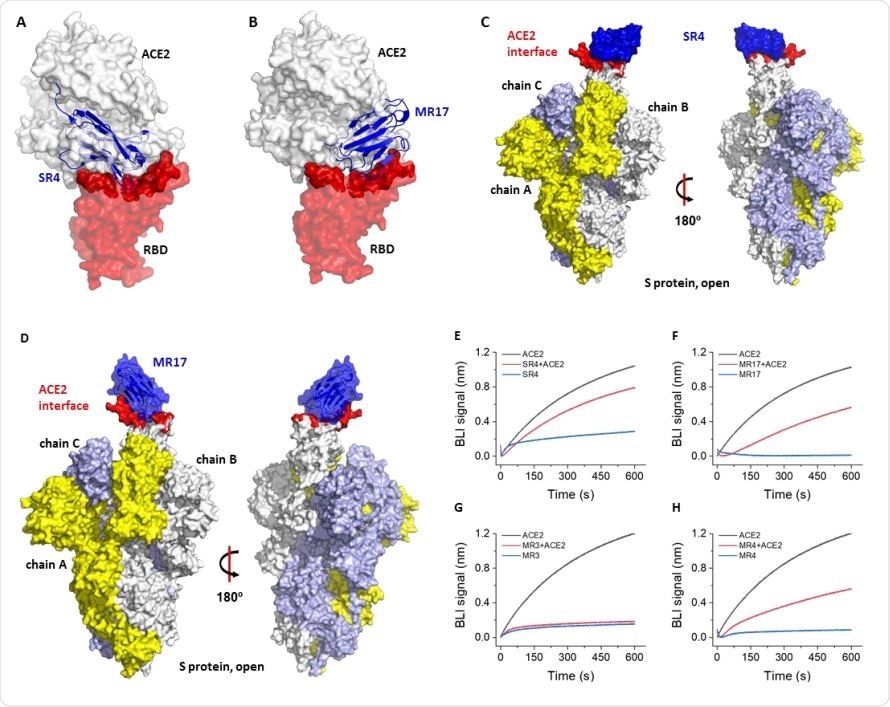
Molecular basis for neutralization. (A) Alignment of the SR4-RBD structure to the ACE2-RBD structure (PDB ID 6M0J) (11) reveals that SR4 (blue) binds RBD (red) at the ACE2-binding site (dark red). The receptor ACE2 is shown as a white surface. (B) Alignment of the SR4-RBD structure to the ACE2-RBD structure (PDB ID 6M0J) (11) reveals that MR17 binds the RBD at the ACE2-binding site. The color-coding is the same as in A. (C,D) Alignment of the SR4-RBD (C) and MR17-RBD (D) to the ‘up’ conformation of the RBD from the cryo-EM structure of the full-length S protein (PDB ID 6VYB) (7). (E-H) Competitive binding for the RBD between sybody and ACE2. A sensor coated with streptavidin was saturated with 1 μM of biotinylated RBD. The sensor was then soaked in 50 nM of sybody with (red) or without 25 nM of ACE2 (blue) for bio-layer interferometry (BLI) assays. As a control, the ACE2-RBD interaction was monitored using sensors without sybody incubation (black).
Crystallization studies narrowed sybody candidates down to two
X-ray crystallography studies performed to elucidate the structure of the candidate RBD-sybody complexes, eliminated MR4-RBD and MR3-RBD, meaning the focus was shifted to SR4-RBD and MR17-RBD.
Both SR4 and MR17 bind to the “seat” and “backrest” regions of the RBD, but neither induced any conformational changes of the RBD, except for MR17 inducing a small “flip” at the backrest region.
Since the study was designed to select sybodies against the RBD of S protein, the researchers further examined the crystal structures to test whether the sybodies suppress viral infection by competing with ACE2 for RBD-binding.
“Superposing the structure of the sybody-RBD complexes to the RBD-ACE2 revealed that they both bind the RBD at the interface where the receptor ACE2 binds,” confirms the team.
A mutant version of MR17 yielded remarkable neutralization efficiency
Further analysis of the structures also enabled the design of an improved version of MR17.
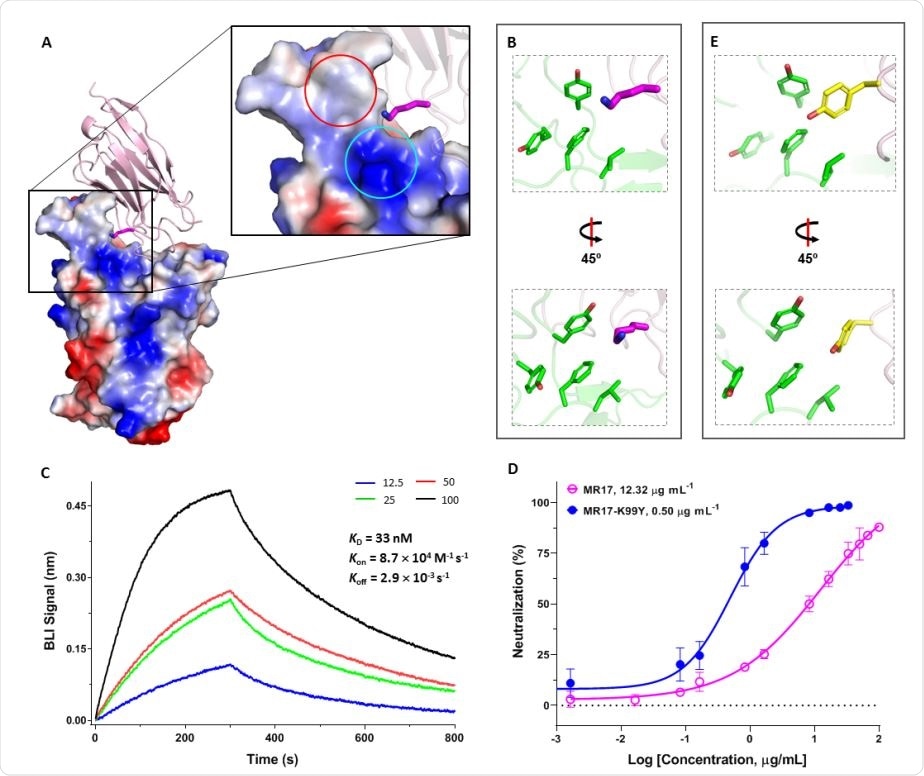
Rational design of a sybody mutant with increased RBD binding affinity and higher neutralizing activity. (A) The overall structure and the expanded view of MR17 (pink cartoon) in complex with the RBD (electrostatic surface). Lys99 on CDR3 (magenta carbon atoms) is poking to an overall positively charged area (cyan circle) which is also adjacent to a hydrophobic patch (red circle). (B) Lys99 from MR17 (magenta) is situated in a hydrophobic patch. (C) Binding kinetics between MR17-K99Y and the RBD. Biotinylated RBD immobilized on a streptavidin-coated sensor was titrated with various concentrations (nM) of MR17-K99Y as indicated. Data were fitted using the built-in software Data Analysis 10.0 with a 1:1 stoichiometry. (D) Neutralization assay of MR17-K99Y (closed circle) using SARS-CoV-2 pseudovirus shows improved neutralizing efficiency compared to the MR17 wild-type (open circle). Data are from three independent experiments. (E) Crystal structure of MR17-K99Y shows that Tyr99 matches the hydrophobic microenvironment.
The team found that the neutralizing ability of MR17 was modest and set out to improve it by designing 19 mutant versions, based on the MR17-RBD structure. This identified a single mutation called K99Y that increased the binding affinity by 1.5 times and the neutralization efficiency by 24 times, compared with wild-type MR17.
“We have generated potent synthetic nanobodies which neutralize SARS-CoV-2 pseudoviruses efficiently and selectively,” said Li and colleagues, who add that the results could pave the way for the development of therapeutic nanobodies to fight COVID-19.
“We anticipate the selection strategy be useful to quickly respond to similar crises should they arise in the future,” the team concludes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources