My interest in protein-DNA interactions and genome editing started a while ago when I collaborated with a biotech company called Cellectis. We performed biophysical studies and solved the crystal structures of homing endonucleases.
These enzymes were one of the first protein scaffolds envisioned as molecular scissors for genome editing. We were one of the first to show that engineered homing endonucleases with new DNA binding specificities were able to correct mutations in human cells involved in diseases such as Xeroderma pigmentosum (Redondo et al Nature 2008).
The finding of CRISPR and the discovery that a simple exchange of the sequence of the associated RNA molecule could direct its cut to the desired sequence was a revolution in the field. So, we started to get interested in CRISPR and its fascinating biology.
Image Credit: Nathan Devery/Shutterstock.com
What are CRISPR systems used for within the scientific industry?
The uses are multiple, ranging from genome editing applications to correct mutations ex-vivo in monogenic diseases to diagnostics of for example cancer or COVID-19, to the possibility to use them as a trigger to deliver drugs contained in hydrogels in a precise time and place. The number of possible applications is growing everyday and many applications have not been discovered yet.
Can you describe how you mapped out and analyzed the Cmr-β complex?
We isolated the complex and used a technique called cryo-electron microscopy (cryoEM) to obtain maps of the coulombic density of the atoms forming the molecule.
We used those maps to build the models of the ribonucleoprotein particle.

How was CryoEM used within your research?
As previously mentioned we obtained maps of the density of the molecule by averaging hundreds of thousands of images of the complex using a method called single-particle averaging.
These maps allow us to identify where the amino acids and nucleotides composing the protein and nucleic acids are organized in the 3D space. Using computer model building programs, we construct the models that help us understand how the different activities of the complex are coordinated.
Why is the Cmr-β complex so complicated to analyze and map?
This has been really difficult work, I would say one of the most complicated in my career, as the complex is around 1 MDa of molecular weight.
It contains 37 different polypeptides and nucleic acids and 6 different catalytic centers: 4 of them cleave the target RNA that hybridizes with the crRNA of the complex, one center synthesizes a cyclic tetra oligoadenylate molecular which activates ancillary RNases and the final one cleaves ssDNA interspecifically.
To find the allosteric coordination between them has required many biochemical studies.
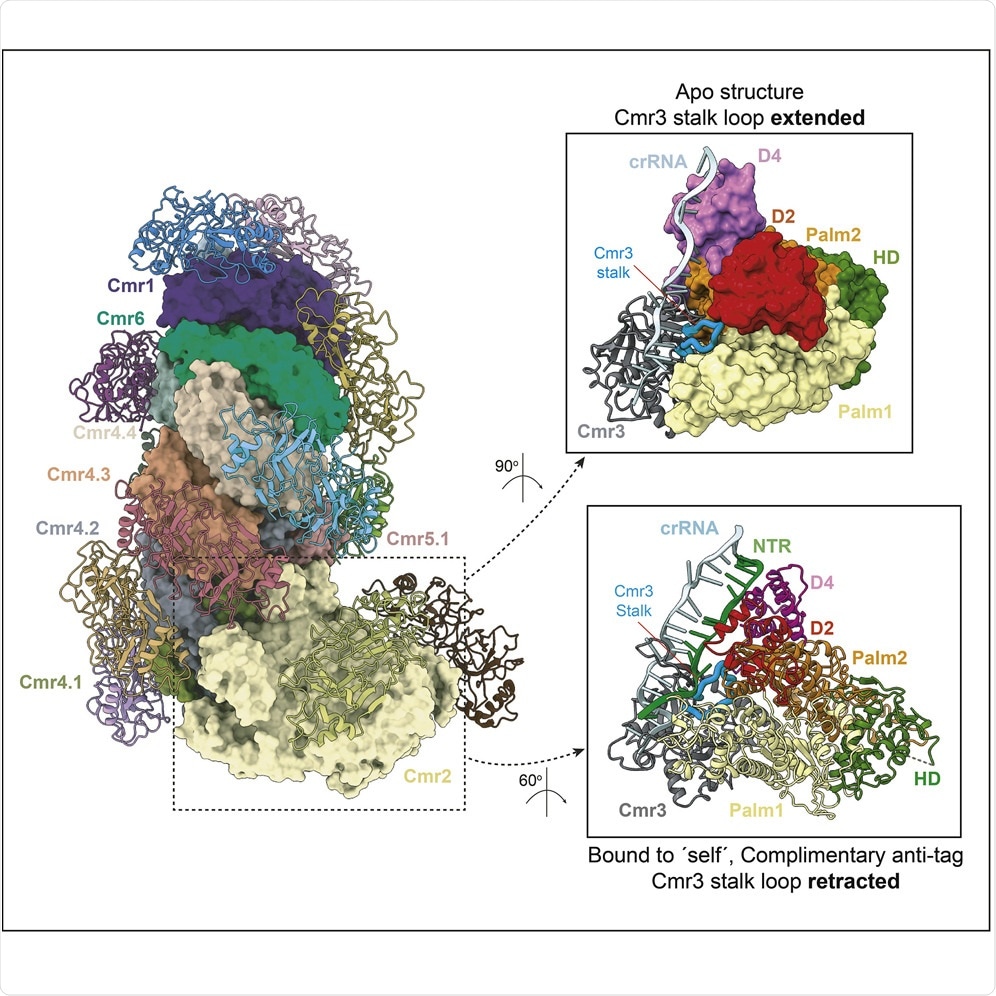
After mapping this complex, what did you discover about its role concerning immune response?
We know now that Cmr-beta has a different mechanism to recognize the invading RNAs from the self-RNA and trigger the immune response.
In addition, we suspect that one of the subunits, Cmr7, may work as a "shield" to protect the complex from the action of anti-crisprs, which are small proteins that can deactivate the effector complex by binding at key places in the effecto complex.
What potential applications could this complex have?
We do not know yet, I do not foresee applications for this system in editing, but I do not discard that it can be tuned for possible biotech applications. In addition, we have filled a gap of fundamental knowledge about Type III CRISPR-Cas effector complexes.
Basic science is essential to bring information that helps us to understand how things work. Later, once this understanding is complete, we can start to redesign nature and think about applications. This is an important concept, for example today we use PCR (polymerase chain reaction) to detect COVID-19. This would have been imposible without previous studies in thermostable polymerases, that open the way to PCR. Kary Mullis was awarded the Nobel prize for its dicovery.
How could further investigation of this complex help in the fight of antibiotic resistance?
This is one of the aspects where I think that our discovery may have future applications as in many cases antibiotic resistance is due to mobile genetic elements (phages and plasmids) that carry resistance genes
It can be that the use of these complexes, after their redesign, can be used to fight resistant bacteria.
Image Credit: Kateryna Kon/Shutterstock.com
What are your next steps in this research?
We will try to understand why there is a combination of different CRISPR systems in some bacteria and how they coordinate the immune response.
Where can readers find more information?
About Professor Guillermo Montoya
Guillermo Montoya is Full-Professor at the University of Copenhagen (UCPH) and Research Director of the Protein Structure & Function Programme at the Novo Nordisk Foundation Centre for Protein Research (http://www.cpr.ku.dk/) since 2014. Montoya is also the Chairman of ISBUC, the Integrative Structural Biology Cluster at UCPH (http://isbuc.ku.dk/).
In the last years, his main research interests are the structural analysis of macromolecular complexes, such as the mammalian chaperonin CCT complex, where he uses a combination of X-ray crystallography and electron microscopy to gain insight about their working mechanisms. Dr. Montoya is also systematically pursuing the structure-function analysis of endonucleases, which are of great interest because of their potential applications in gene therapy.
Recently, he solved the structure of a new CRISPR-Cas member, Cpf1, which can open new avenues in genome modification. He is a member of international project evaluation panels and also reviews projects for European funding agencies (ERC, EU, SNF, DFG, ANR, NOW, MINECO, Welcome Trust, Vinnova) and serves as an associate editor and ad-hoc reviewer for scientific journals. Montoya was elected EMBO member in 2018 and received the Novo Nordisk Distinguished Investigator Grant in April 2019.