Understanding the invasion process of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the host cell is crucial in therapeutic designs. Depending on the approach, it also throws light on the evolution of the virus and its infection mechanism. The proteolytic activation of the SARS-CoV-2 spike protein, involved in host cell entry, is evaluated in detail in this study.
The SARS-CoV-2 virus that causes the disease COVID-19 (coronavirus disease) has now infected more than 35.65 million individuals and claimed over 1 million lives worldwide. The rise of the pathogenic strains of human coronaviruses (CoVs), with the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, the Middle East respiratory syndrome coronavirus (MERS-CoV) first emerged in 2012.
It is essential to understand the host-virus interaction and the sequential processes that ensue towards an infection.
In a recent paper published on the bioRxiv* preprint server, Cornell University researchers study the proteolytic activation of the SARS-CoV-2 spike sites, re-evaluating the furin cleavage. Their results demonstrate that S1/S2 pre-cleavage is essential for plasma membrane entry into Calu-3 cells (a model lung epithelial cell line), but not for endosomal entry in Vero E6 cells (a model cell culture line).
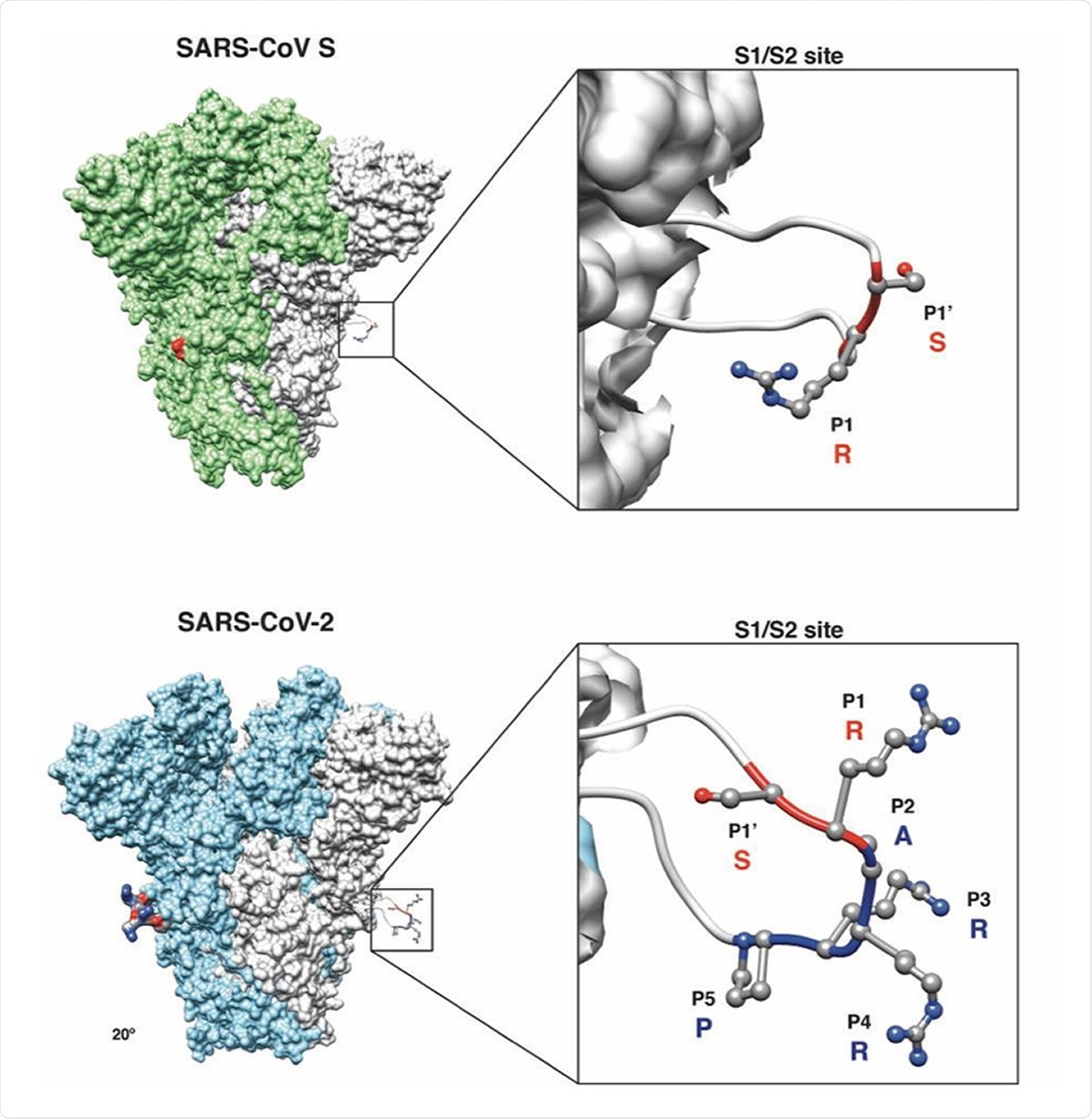
Predicted structural model of the SARS-CoV and SARS-CoV-2 S proteins. (Inset) Magnification of S1/S2 site with conserved R and S residues (red ribbon) and the unique four amino acid insertion P-R-R-A for SARS-CoV-2 (blue ribbon) are shown. The P’s denote the position of that amino acid from the S1/S2 cleavage site, with P1-P5 referring to amino acids before the cleavage site and P1’ referring to amino acids after the cleavage site.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study reports that apart from furin, other proteases are also responsible for processing SARS-CoV-2 S1/S2.
The spike (S) glycoprotein of SARS-CoV-2 is used to mediate viral entry into host cells. It is a large transmembrane protein that decorates the virus particle.
It is known that the S protein cleaves at the S1/S2 or S2’ site. This is essential for viral entry, occurring at either the cell plasma membrane or the endosomal membrane. It is known that the S1/S2 site of the spike protein can be cleaved by furin - this expands viral tropism to lung cells. The viral tropism is the ability of different viral strains or isolates to infect different cell types or tissues, inducing virus production as a result of infection.
The results of the initial viral infection can vary widely depending on the site and way of entry, the cell types infected, the responses of local immune cells, and the virus or host species. In this study, the authors analyze the furin S1/S2 site in the related CoVs.
Based on the binding strength and accessibility of the motif in the furin binding pocket, the authors analyzed the likelihood of S1/S2 site cleavage by furin, using the prediction tools PiTou and ProP. Though both algorithms agree with each other in predicting furin cleavage, due to its better sensitivity and specificity, they use the PiTou algorithm for further analysis. The algorithm predicts that the SARS-CoV-2 S1/S2 site can be cleaved by furin, whereas the other proposed lineage precursors cannot be cleaved by furin - this is experimentally confirmed. Also, the SARS-CoV-2 S1/S2 site has increased furin recognition compared to the other B-betaCoVs; the site is also not optimal for furin cleavage. The authors discuss in detail the origin and evolution of the virus and the related significance of the cleavage site. They urge to explore alternative explanations for the SARS-CoV-2 S1/S2 site as a gained furin insert.
In this study, the authors used viral pseudoparticles to assess the functional importance of the S1/S2 site for SARS-CoV-2 entry. The core of the pseudoparticle is a murine leukemia virus (MLV), and the viral envelope protein is decorated to recapitulate the entry steps of their native counterpart accurately. These particles contain luciferase reporter - that integrates into the host cell genome and produces quantifiable luciferase upon successful infection. This study confirmed that the envelope protein is driving infection. They observed higher infectivity of SARS-CoV compared to SARS-CoV-2 - it is from more S protein incorporated in the former.
The study suggests that the S1/S2 cleavage is essential for TMPRSS2 mediated plasma membrane entry, but not for cathepsin L mediated endosomal entry and that furin may not be the only protease responsible for the S1/S2 cleavage event.
The claimed SARS-COV-2 S1/S2 site is essential for the virus to enter the plasma membrane in respiratory cells; without the cleavage, the virus would be endocytosed, and it will not be effective in infection. It is also known that this site activation appears to have a role in the immune response against SARS-CoV-2. The authors show that other proteases also need to be evaluated. The overall observations from the study support that the role of the SARS-CoV-2 S1/S2 site is to expand viral tropism to lung cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Proteolytic activation of the SARS-CoV-2 spike S1/S2 site: a re-evaluation of furin cleavage, Tiffany Tang, Javier A. Jaimes, Miya K. Bidon, Marco R. Straus, Susan Daniel, Gary R. Whittaker, bioRxiv 2020.10.04.325522; doi: https://doi.org/10.1101/2020.10.04.325522
- Peer reviewed and published scientific report.
Tang, Tiffany, Javier A. Jaimes, Miya K. Bidon, Marco R. Straus, Susan Daniel, and Gary R. Whittaker. 2021. “Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin.” ACS Infectious Diseases 7 (2): 264–72. https://doi.org/10.1021/acsinfecdis.0c00701. https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00701.