The COVID-19 pandemic is caused by the zoonotic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Like other pathogenic coronaviruses, SARS-CoV-2 crossed the species barrier to cause human disease. This fact motivated the current study, published on the preprint server bioRxiv* in October 2020, which explores the way coronaviruses engage their host cell receptors in a range of animals.
This study's importance lies in the close interactions between humans and domestic animals of various sorts, where the respiratory spread is very likely to occur. Not only is this a constant threat to the human race, but it poses an economic challenge to livestock farmers and pet owners.
Studies on viral pathology and to measure candidate therapeutic drugs also use animals infected with the virus. Thus, this study will assist public health experts, economic strategists, and those working with animal models of COVID-19.
Viral Binding Via Spike Protein
The virus binds to the angiotensin-converting enzyme 2 (ACE2) on the host cells in the lungs and other organs at high affinity. The result is the virus's fusion to the host cell membrane, which leads to cell entry and infection.
The SARS-CoV-2 spike-ACE2 complex has been studied crystallographically and by electron microscopy. This showed how the amino acids interacted at the spike receptor-binding domain (RBD) and the human ACE2.
Being able to measure potential spike RBD binding would help to be able to predict which animals are susceptible to this virus.
Variability in the ACE2 Sequence
The researchers in the current study examined the conserved and divergent sequences of the ACE2 receptor protein in a number of relevant species, especially in the receptor-binding domain (RBD).
All vertebrates had homologous ACE2 sequences to human ACE2. The sequences from cynomolgus macaque and rhesus monkeys were 95% and 94% identical to human sequences. For other mammals, it was in the order of 80-87%.
With the horseshoe bat, suspected to be a reservoir for the virus, the identity was only 81% for the whole ACE2 protein and 76% for the 25 interacting amino acids. In fact, in many cases, the degree of identity was widely different across the whole sequence and the interacting residues.
Corresponding with this finding, they observed that one ACE2 protein, from cattle (Bos taurus), though different from the human variant across the whole sequence (79% identity), showed high-affinity binding to the SARS-CoV-2 spike RBD (84% identity). This suggests that there may be many species of mammals that allow infection with this virus.
On the other hand, binding site residues are lower in identity relative to the whole sequence in dogs and rabbits. Future studies must establish the hypothesis that the residues that interact with the RBD are much more relevant to binding than those outside this site.
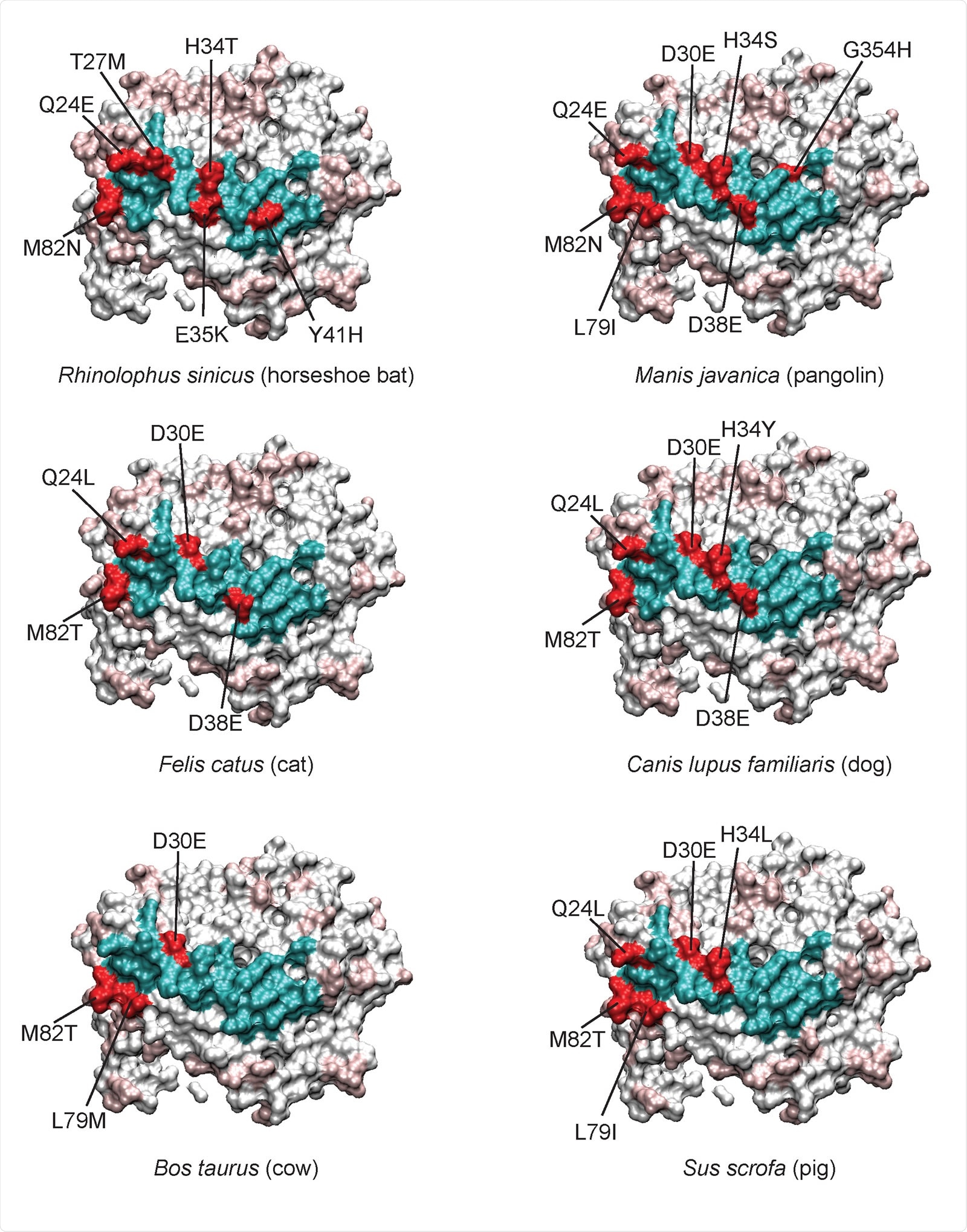
Variable residues at the ACE2-RBD interface of individual species. With human ACE2 360 as a reference, the variant interacting residues for each species are colored red, and conserved 361 residues colored cyan. For the remainder of the ACE2 protein, conserved residues are white and 362 variable residues light red. Certain residues like M82, Q24, D30, and H34 are often mutated relative to 363 human. D30 is conservatively changed to glutamate, however more non-conservative changes can be 364 seen for H34, for example.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
ACE2 Sequence and Binding Affinity
The researchers compared the importance of various regions and of individual amino acids of the protein to its structure. This type of study has earlier shown the presence of complementary determining regions (CDR) within antibodies, which interact with the corresponding antigen.
A powerful tool used for variability analysis is Shannon entropy. Such tests showed the ACE2 to be highly conserved across the whole sequence. In the 25 RBD-binding or interacting residues, 21 were well conserved, and none had high Shannon entropy.
There were 5 fully conserved residues, 4 being near the binding site, and only one being a contact residue. Their preservation indicates the importance of these to cell entry via protease-mediated degradation or to the protein structure among species.
The most variable were at positions 24 and 34, the latter being in the middle of the ACE2-RBD interface and having the highest Shannon entropy by far. Both of these are contact residues. Some investigators have suggested that these positions may help predict the infectivity of a given SARS-CoV-2 variant.
Matching Residues and Binding Affinity
The residues that failed to match human ACE2 were evaluated separately in the next step. They found that with the horseshoe bat, only 5 of 8 residues in contact with the RBD were the same as in human ACE2, but 19/25 nearby residues. This could mean that its ACE2 has a lower affinity for the virus.
Another putative intermediate animal host for the virus was the pangolin, and here only one contact and three adjacent residues failed to match that of human ACE2. More data is required to confirm its role in SARS-CoV-2 transmission. Similar close matches include dogs and cats.
In cattle, three interacting residues at the binding site are altered, one being a contact residue. Cows are known to harbor a distantly SARS-CoV-2-related bovine coronavirus, and two coronaviruses in this family have jumped across the species barrier already to infect humans with the common cold.
However, these bind to sialoglycans rather than ACE2. Nonetheless, it is necessary to evaluate cows for their ability to act as a reservoir host for SARS-CoV-2. In fact, interactions between bovine coronavirus and SARS-CoV-2 could lead to recombinants emerging, with dangerous consequences.
Bovine ACE2 Highly Conserved at Binding Site
While the ACE2 of cows is only about 78% identical to humans, the residues at the binding site show higher (84%) conservation. The researchers examined if this would allow high-affinity binding to the target virus RBD despite the rest of the ACE2 sequence's lower homology.
They found that on ELISA testing, the binding of bovine ACE2 appeared to be ten times lower than for human ACE2, mostly because of a more rapid rate of dissociation from the complex once formed. Surprisingly, this lower affinity was comparable to that of human ACE2 for SARS-CoV-1 RBD, being in the low nanomolar range compared to the latter's sub-nanomolar concentration.
Best Fit with Two-Site Binding Model
Both human and bovine ACE2 showed a much more consistent fit when a two-site model was used to simulate the interaction with the RBD of the virus. This may mean that RBD multimers are formed to bind avidly to the surface of the ACE2. If so, the presence of a second site would explain the tenfold higher affinity.
Implications
The researchers comment, "Such interactions would be important to explore to understand the details of the interaction between coronavirus spike RBD and ACE2 on the cell surface." Bovine ACE2 binds the SARS-CoV-2 RBD with high affinity, even though it is 5-10 times lower than with human ACE2, and appears capable of allowing the virus to infect cows.
The study also shows the feasibility of using the receptor's biochemical characterization to predict how various species will treat the virus, especially with high-speed screening.
The researchers indicate, "Further study into various species' viral receptors, including the recently discovered neuropilin-2 co-receptor for SARS-CoV-2, should enable much more accurate and rapid development of predictive algorithms based on viral receptor sequence and structural modeling."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources