A team of scientists from the University of Oxford, UK, has revealed that site-specific glycosylation differs between spike proteins derived from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and a viral vector-based vaccine candidate. A significantly high amount of non-physiological glycosylation may impair the ability of a vaccine candidate to induce desired immune responses. The study is currently available on the bioRxiv* preprint server.
SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19), is a single-stranded, positive-sense RNA virus with a genome size of about 30 kb. The virus spread rapidly from person to person, primarily via respiratory droplets. In addition to implementing non-pharmacological control measures (wearing masks, washing/sanitizing hands, and movement restrictions), several attempts have been made to develop effective therapeutics and vaccines. Although many potential vaccine candidates are currently in the pipeline, it is still uncertain how effective these vaccines are in terms of inducing and maintaining SARS-CoV-2 specific immune responses.
Mechanistically, the interaction between the viral spike protein and host angiotensin-converting enzyme 2 (ACE2) facilitates the entry of SARS-CoV-2 into host cells. The spike protein is a viral surface protein that undergoes N-linked glycosylation (addition of N-glycans to 22 N-glycosylation sites) after its synthesis in the host cell endoplasmic reticulum. The addition of O-glycans occurs in the Golgi.
The glycosylation of spike protein is essential for maintaining viral protein stability, influencing viral infectivity, and facilitating immune evasion. Inside host cells, viral spikes are the most frequently targeted molecules by neutralizing antibodies. Thus, most of the vaccine candidates are designed using viral spike protein as an antigen to induce desired immune responses. However, both N-linked and O-linked glycans on the spike surface protect underlying viral epitopes from the attack by host neutralizing antibodies. Sometimes, these glycans act as viral epitopes to trigger host cell immune responses. Therefore, proper knowledge about the spike protein glycosylation process is essential for developing an effective vaccine candidate.
Current study design
The scientists aimed to compare the spike protein's glycosylation process derived from SARS-CoV-2 and a viral vector-based vaccine candidate. They used lung epithelial cells to grow SARS-CoV-2 and subsequently used an anti-SARS-CoV-2 monoclonal antibody to purify the spike protein.
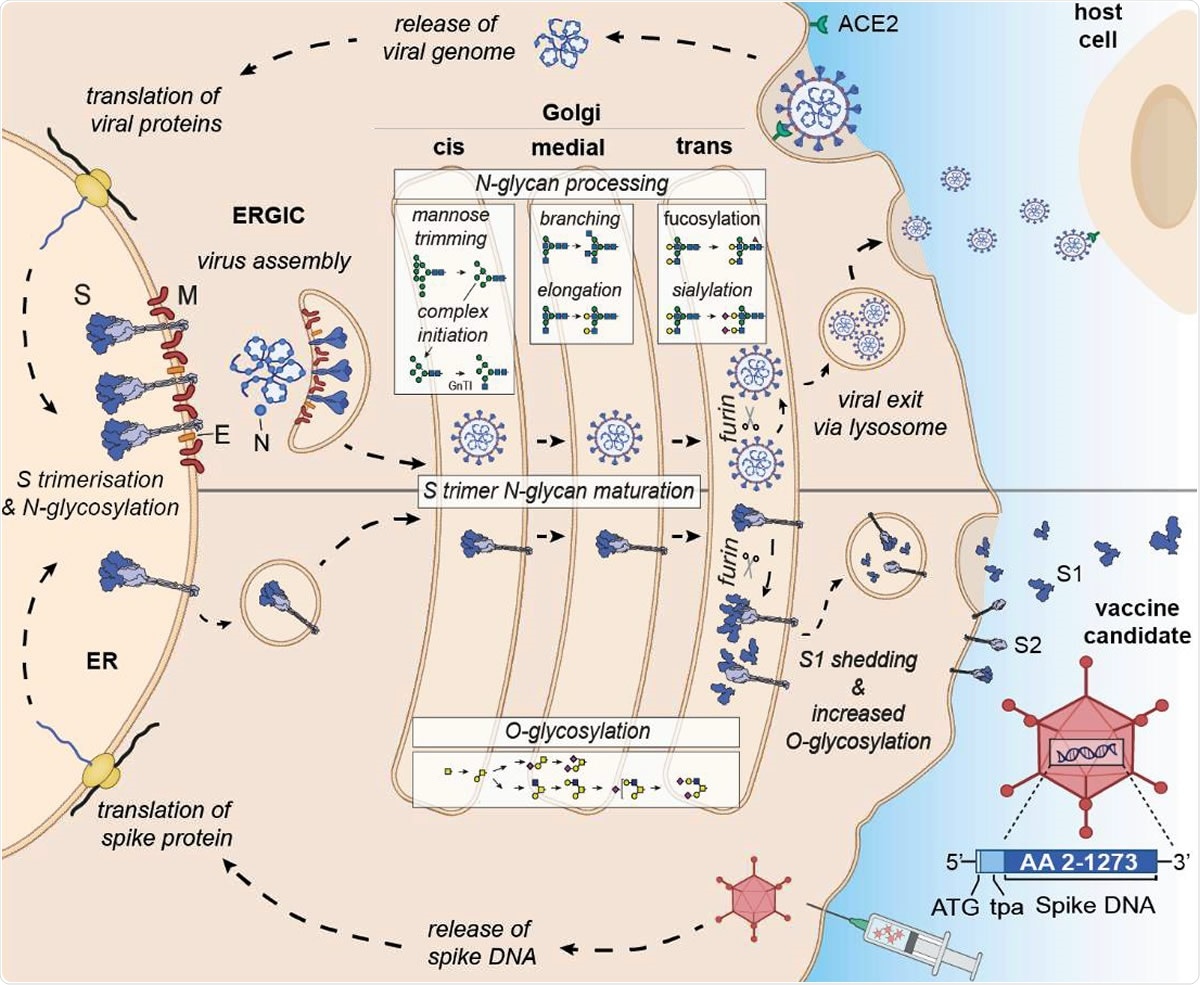
Differential expression and glycan processing of virions and vaccine-derived spike glycoproteins. SARS-CoV-2 binds to its receptor ACE-2 and infects cells, leading to the release of the viral genome and translation of viral proteins. Spike protein is co-translationally N-glycosylated and forms trimers in the ER that traffic to the ERGIC where they are incorporated into budding virions. Individual virions continue through the secretory pathway to the trans-Golgi prior to following a lysosomal egress route. For the vaccine candidate, spike DNA is administered via an adenovirus vector system, and spike protein is synthesized in the ER, where it is N-glycosylated and trimerizes as before, but as it is not incorporated into a budding virion in the ERGIC, it continues through the secretory pathway and, via lysosomes, to the plasma membrane. In both cases, the spike glycoproteins have access to both the N- and O- linked host glycosylation machinery. Upon furin cleavage in the trans-Golgi, S1 and S2 of the virus stay non-covalently associated, whereas furin cleavage of the vaccine antigen results in shedding of monomeric S1vaccine antigen. Glycomic signature analysis of these two proteins shows that the N-linked glycosylation occupancy levels, which are determined in the ER, are comparable for S1virus and S1vaccine antigen whereas the attached glycoforms vary reflecting their different accessibility to glycan processing enzymes. S1vaccine antigen carries not only higher levels of complex N-glycans but is also extensively O-glycosylated after furin cleavage in the trans-Golgi, when most S1vaccine antigen is shed and secreted in a soluble monomeric form. Some S1 and S2vaccine antigen is displayed on the cell surface, presumably as trimers.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Important observations
The ultra-performance liquid chromatography-based analysis of virus-derived N-glycans exhibited 79% of complex-type N-glycans and 21% of oligomannose/hybrid N-glycans. In contrast, analysis of vaccine-derived N-glycans exhibited 89% of complex-type N-glucans and only 11% of oligomannose/hybrid N-glycans. These findings indicate that the glycan processing of SARS-CoV-2-derived spike protein differs significantly from that of the vaccine-derived spike protein.
The scientists conducted mass spectrometric analyses to evaluate the spike protein glycan processing further. They observed that N-glycan processing of the spike protein's S1 subunit is comparable between the virus and the vaccine candidate. However, they observed O-linked glycosylation at the T678 site on the virus-derived spike protein, which was absent on the vaccine-derived spike protein. This finding indicates that the viral spike protein maintains a more flexible configuration than the vaccine-derived spike protein.
Furthermore, the scientists expressed spike protein (similar to vaccine candidate) in mammalian cells and compared its glycosylation process with the virus-derived spike protein. Interestingly, the S1 subunit of expressed spike protein displayed 96% of complex-type N-glycans and only 4% of oligomannose-type N-glycans. This indicates that compared to the S1 subunit of viral spike protein, the S1 subunit of expressed spike protein is extensively processed by glycosylation enzymes.
They observed that extensive N-glycan processing is prevented at the N234 site because of the spike protein's spatial and temporal assembly in the host endoplasmic reticulum and Golgi. For both vaccine-derived and virus-derived S1 subunits, this site remained fully under-processed (100% oligomannose); however, for expressed protein-derived S1 subunit, some level of glycan processing (75% oligomannose) was observed at this site. With further analysis, the scientists observed that the dissociation between S1 and S2 subunits of the expressed spike protein occurs in the trans-Golgi and not in the cellular plasma membrane. By individually expressing recombinant S1 subunit that cannot trimerize, they observed 100% complex-type N-glycans at the N234 site and 100% O-glycan at the T678 site.
Taken together, the study findings indicate that glycan processing of viral proteins should be critically reviewed before developing vaccine candidates against SARS-CoV-2. A significantly high amount of complex N-glycans can potentially cover viral epitopes, which in turn can inhibit the antibody-epitope interaction and prevent the induction of desired immune responses. According to the scientists, a vaccine candidate containing prefusion-stabilized spike protein without a proteolytic cleavage site is optimal for inducing strong and sustained immune responses. Inhibition of S1 subunit shedding by abolishing the proteolytic cleavage site is important for a vaccine candidate's proper immunogen presentation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources