The reported early success of several coronavirus disease 2019 (COVID-19) vaccines undergoing clinical trials at present has grabbed the world's attention.
Image of the coronavirus spike protein that mediates coronavirus entry into host cell. 3d illustration. Image Credit: Design_Cells / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Transition to endemicity
The SARS-CoV-2 virus is a betacoronavirus, belonging to the coronavirus (CoV) family, including both SARS-CoV and MERS-CoV. These are RNA viruses with very large genomes, about 30 kb in length. The family gets its name from the characteristic 'crown' of spike proteins protruding from the virus's surface.
The spike glycoprotein is the primary viral antigen involved in viral entry into the host target cell. It is also the prime target for most vaccines being developed at present.
If vaccines are unable to induce protective immunity, the COVID-19 pandemic will slowly subside over time to become an endemic illness, such as influenza. This latter family of viruses has shown the same pattern at least four times over the last 100 years, as have seasonal CoVs like OC43.
This transition is marked by the emergence of mutations in the viral genome, resulting in an altered protein product and therefore disrupts the binding of specific protective antibodies to the viral antigen. Thus, the endemic strain can evade the immune response elicited by earlier strains of the same virus.
This response is seen with influenza, which has an RNA-dependent RNA polymerase (RdrP) that accumulates numerous errors over the years, finally achieving a very different antigenic profile at the end of this period. However, because of the efficient proofreading apparatus seen with CoV RdRps, the substitution rate of nucleotides is slower relative to other RNA viruses. This has led to the hope that the SARS-CoV-2 spike antigen would remain stable, and that all the currently circulating strains would thus be susceptible to the neutralizing antibodies developed in response to the early vaccines.
Independent recurrent deletions
The current study reports, however, that the virus is adapting to the presence of immunity, as signaled by repeated deletion mutations at specific sites of the spike protein, causing the rapid evolution of spike antigenic diversity. These deletions have been observed both in viral sequences isolated from chronically immunosuppressed patients from all over the world.
The specific deletion sites are termed recurrent deletion regions (RDRs), and all four are in the N-terminal domain (NTD) of the spike glycoprotein. All are at defined antigenic sites. One single variant is present in over 2.5% of all isolates from circulating viruses at present.
This virus is therefore finding niches of antigenic variation that enable it to escape neutralization by preventing the binding of the protective antibody to the altered antigen. The escape mutation is also transmitted to other individuals.
Different patients, common NTD mutations
One patient with cancer contracted the virus, and despite treatment with convalescent plasma and remdesivir, was unable to clear the infection. The isolate showed the spike gene had NTD deletions, termed PLTI1. Similar deletions were found in longitudinal samples from eight patients, in the GISAID database, with six being identical or near-identical to PTLI1. Each of these deletion variants stemmed from an early intact spike sequence. Also, among these patients, seven had individually unique patterns of substitutions over the whole period of sample collection, refuting the hypothesis that the deletions came from other individuals at the hospital or from the community.
Instead, the researchers say, "The most parsimonious explanation is that each deletion arose independently in response to a common and strong selective pressure."
In GISAID, they found over 1,000 viruses with spike deletions, and 90% of these were in the RDRs. Most of these apparently arose during replication. Over 90% were in-frame deletions, consisting of 3 or sets of 3 nucleotides being deleted. The eventual outcome was an open reading frame rather than a stop codon, in over 97% of deletions in the NTD. In other parts of the spike protein, deletions follow this trend only about a third of the time. In other words, RDRs have an inherent tolerance for deletions and accumulate them.
RDRs may show anywhere from a unique single-virus deletion to those which are found in multiple variants. Of the four RDRs, 1 and 3 typically show a single-site predilection, belong to clade G and GR and are mostly from Europe. This indicates a selection advantage for certain deletions in some clades, which are limited to certain regions. Conversely, at the other two, there are a number of overlapping deletions, found in strains from all over the world.
Transmission of deletion variants
For each of the RDRs, identical deletions have been found in different patients simultaneously, indicating that these strains spread between humans. This is probably due to bouts of viral transmission from infected individuals to others.
Most importantly, the presence of RDR variants acts as a type of escape mutation, allowing the virus to escape from a strong selection pressure applied commonly to all variants. RDR1 and 3 are on one side of the spike NTD, while 2 and 4 are on the other side. Both form antibody epitopes, or binding sites. The neutralizing antibody 4A8 is found to be within the connecting loops where RDR 2 and 4 are located, but binding is abolished by three deletions, one in 4 and two in RDR2. However, these deletions still allowed binding by a monoclonal anti-RBD antibody.
Deletions at RDR 1 and 3 allowed binding to occur with either of these antibodies, thus showing that these are on separate sites from the other two. Thus, whether in single RDRs or between pairs of RDRs, the occurrence of recurrent deletions associated with distinct epitopes, leading to the abolition of binding, but allowing the protein to retain its function, shows the change in antigenicity between natural SARS-CoV-2 variants.
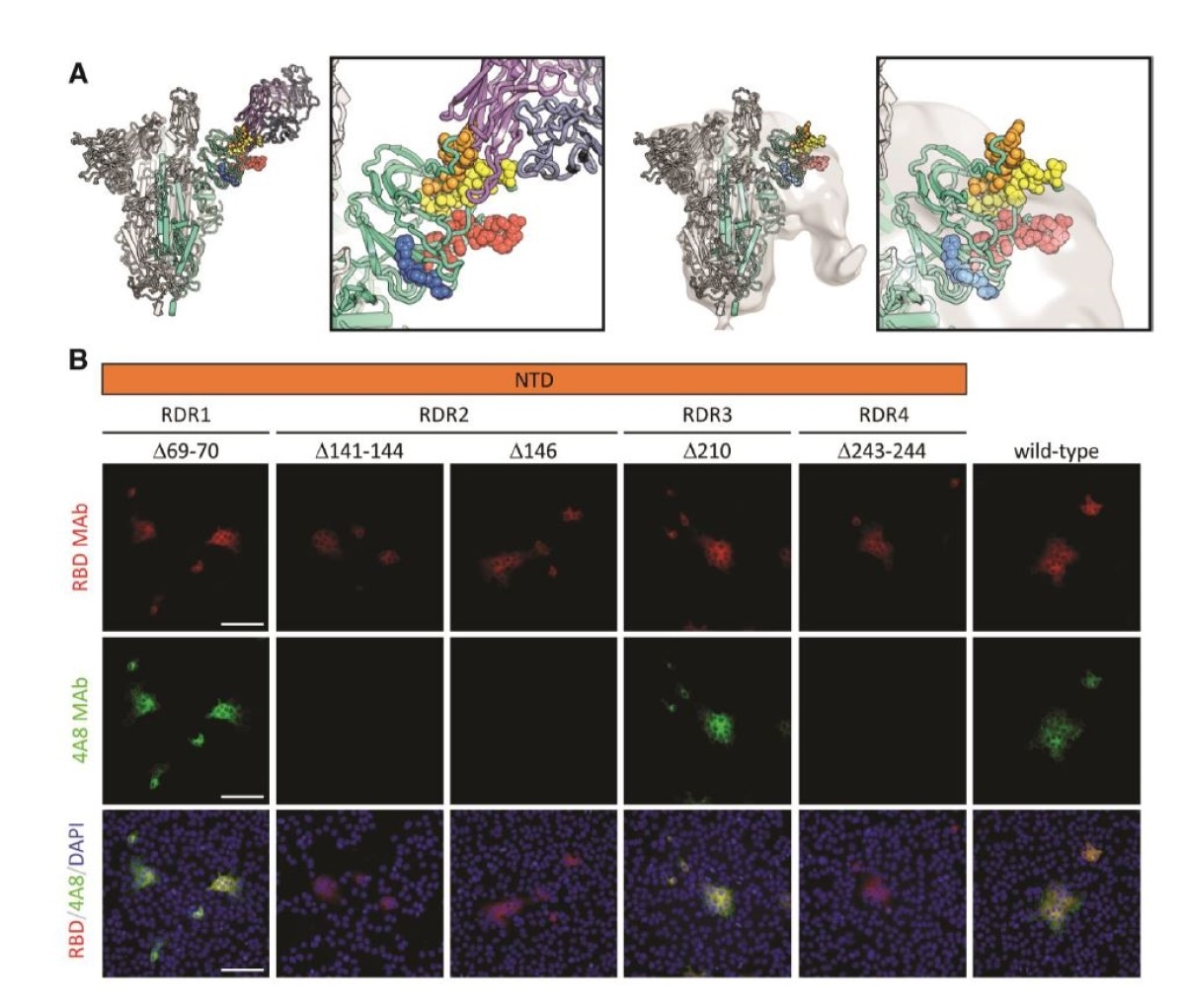
Deletions in the spike NTD alter its antigenicity. RDRs map to defined antigenic sites. A.Top: A structure of antibody 4A8 (16) (PDB: 7C21) (purples) bound to one protomer (green) of aSARS-CoV-2 spike trimer (grays). RDRs 1-4 are colored red, orange, blue, and yellow, respectively, and shown in spheres. The interaction site is shown at right. Bottom: The electron microscopy density of COV57 serum Fabs (20) (EMDB emd_22125) fit to SARS-CoV-2 Spike trimer (PDB: 7C21). The same view of the interaction site is provided at right. B. S protein distribution in Vero E6 cells at 24 h post-transfection with S protein deletion mutants, visualized by immunodetection in permeabilized cells. A monoclonal antibody against SARS-CoV-2 S protein receptor-binding domain 19 (RBD MAb; red) detects all mutant forms of the protein (?69-70, ?141-144, ?146, ?210, ?243-244) and the unmodified protein (wild-type). 4A8 monoclonal antibody (4A8 MAb; green) does not detect mutants containing deletions in RDR2 or RDR4 (?141-144, ?243-244, ?146). Overlay images (RBD/4A8/DAPI) depict co-localization of the antibodies; nuclei were counterstained with DAPI (blue). The scale bars represent 100 µm.
What are the implications?
The researchers point out, "The simplicity of using deletion to drive diversity is biologically compelling." Selection pressure rarely operates during a pandemic, since the infection typically resolves rapidly, even before antibody production is complete.
During the endemic stage, however, the situation is different, since the presence of antibodies in already recovered individuals, and in people who receive passive immunity in the form of convalescent plasma, or therapeutic monoclonal antibodies, exerts selection pressure on the virus. This will then cause viral variants with the ability to evade these antibodies to be selected.
In individuals, several different variants of the same virus can emerge over time, giving rise to quasispecies. This explains how the same deletion can repeatedly appear in many different individuals with long-standing infection. The researchers explain, "SARS-CoV-2 is continuously exploring sequence and antigenic space in different genetic, environmental and geographical contexts." One variant, called Δ69-70, has rapidly become more frequent, forming 2.5% of all sequences in October 2020, up from 0.01% in July 2020.
The change in binding behavior is concerning since many of these deletions are found in strains circulating across the UK, where Phase III trials of a COVID-19 vaccine is ongoing. Such behavior is bound to continue, say the researchers, leading to the endemicity of SARS-CoV-2 infection.
To counter this by up-to-date vaccine development or other strategies, they conclude, "Efforts to track and monitor these recurrent, rapidly arising, geographically widespread variants are vital."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources