The current coronavirus disease 2019 (COVID-19) pandemic has claimed over 1.9 million lives so far, and scientists have been racing to develop vaccines that will curtail its spread. Most of these target the spike (S) protein of the virus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The spike is anchored to the host cell membrane, as it mediates viral binding to the host cell receptor. This is the angiotensin-converting enzyme 2 (ACE2).
The virus's cytosolic tail also interacts with the host cell to promote its entry into the endoplasmic reticulum (ER). A new study appearing on the bioRxiv* preprint server elucidates the conformation of this region of the virus, which will help understand how its form is related to its function in viral entry.
The structure of the spike
The spike protein is located on the surface of the virus and thus presents a prime target for antibodies as well as cell-mediated immune responses. Many vaccines and therapeutic antibodies are also designed on the basis of this glycoprotein, which is a trimer comprising an extracellular unit and a cytoplasmic domain. The former is the anchoring unit because its transmembrane (TM) domain is located in the viral membrane.
The spike is first secreted as a monomer from the endoplasmic reticulum (ER), but then becomes a homotrimer in order to gain entry to the Golgi complex. Within these ER-Golgi compartments, it gains N-linked mannose-rich oligosaccharides as its side chains, which are then further modified. It has two subunits, S1 and S2, that have a non-covalent association in the metastable prefusion state. This spike protein is cleaved by furin or similar proteases at the S1/S2 cleavage site.
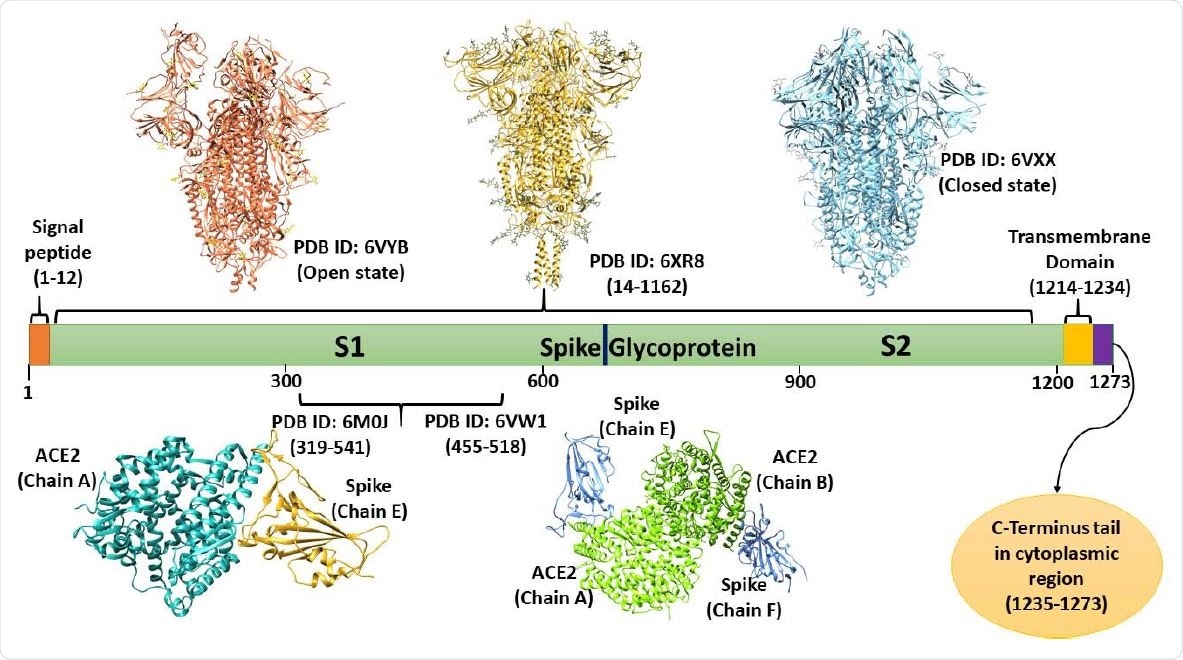
Domain architecture of spike Glycoprotein: depiction of available structures in open and closed states, transmembrane domain, and cytoplasmic C-terminal tail (based on UniProt database).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The S1 subunit has an N-terminal domain, a C-terminal domain, and two other subdomains. The receptor-binding domain, or RBD, is synonymous with the C-terminal domain and contains the receptor-binding motif (RBM). The RBM binds to the ACE2 receptor, being an extended loop insertion. Once attached to the host cell via the ACE2 receptor, the S2 subunit mediates the fusion of the host cell and the viral membrane.
The cytoplasmic tail is still not well known but contains an ER retrieval signal that is shared by other similar viruses. A novel dibasic motif quite similar to the above is found at the two ends of the C-terminal domain (CTD), and is essential for its localization within the cell. In some cases, deletions of the CTD have been associated with increased infectivity in some pseudoviruses carrying the spike protein.
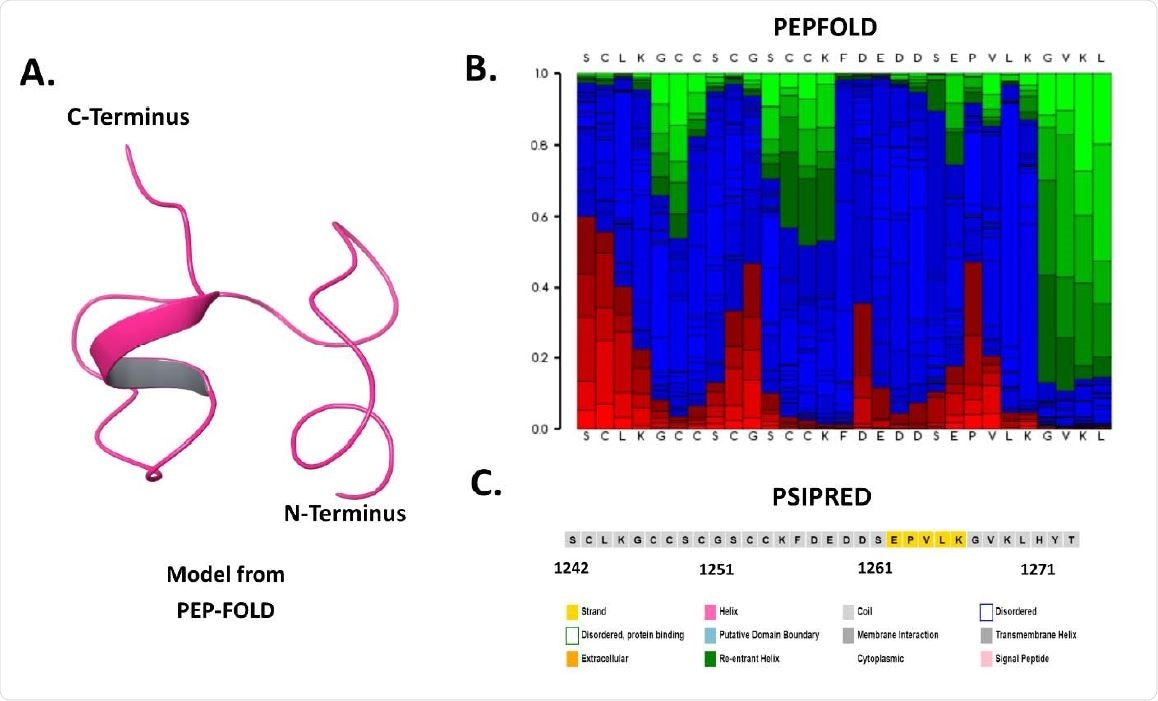
Sequence and structure-based analysis of spike C-terminal cytoplasmic domain (1242-1273 residues): A. Modeled structure through PEP-FOLD web server, B. Secondary structure analysis using PSIPRED web server, and C. PEP-FOLD structure analysis depicting helix (red), coil (blue), and extended (green).
Cytoplasmic domain is a molecular recognition feature
The same researchers who carried out the current study have already shown that the cytoplasmic tail is a MoRF (Molecular Recognition Feature) region. These regions are parts of a protein that are disorder-based binding regions, participating in binding other proteins and DNA or RNA. The researchers aimed to explore the cytoplasmic domain from residues 1242-1273 of SARS-CoV-2. They looked at the molecular dynamic (MD) simulations up to one microsecond (μs) to elucidate the structure-function relationship of this domain, which would be helpful to understand how the spike protein interacts with other proteins, both viral and host.
Transmembrane analysis
Computational approaches were used to study the secondary and tertiary protein structure. MD simulations can further show protein structural conformations at the level of atoms. The researchers also analyzed the sequences of the transmembrane region and disorder prone regions. The predictors used in this study showed that the spike TM region is within 1213-1246 residues. The transmembrane region from residues 1216-1241 has already been reliably predicted by a consensus-based server named CCTOP that compares available experimental evidence of closely related proteins.
The current study thus focused on the region from 1242 to 1273 residues. Since this protein structure has not been determined experimentally, an approximate structure was obtained by an approach called structure modeling. This uses many optimized algorithms to understand the homology and properties of amino acids in the structure. Once a preliminary structure is ready, the fragmented prototypes are assembled using the optimized potential for efficient structure prediction (OPEP), a forcefield simulation at medium scale.
Disorder in cytoplasmic region
After obtaining the best structure of the cytoplasmic tail's tip, the structure was prepared to carry out more MD simulations under aqueous conditions. This structure has two small helical regions, which contribute over 12% of the secondary structure, the rest is made up of turns and long coils. Throughout the simulation, the cytoplasmic tail region remained unstructured, showing it to be a disorderly region.
Structure gain with rising temperature
This region proved to have a secondary structure, a β-sheet conformation at the N-terminal end, containing two beta-strands that have regular variations of amino acid length once every 0.1 μs frame unless they are 0.3 μs and 0.4 μs frames that lack secondary structure. Once a 0.5 μs frame is achieved, beta strands begin to be slowly lost, but the spike C-terminus shows a gain in disorder. Therefore, “the 1 μs simulation frame comprises of only a short β-strand and rest unstructured residues.”
These changes in the structure have led to considerable fluctuations in the atomic distance throughout the MD simulation. Along with changes in the composition of the secondary structure, structural inconsistency is also apparent throughout. Thus, the spike cytosolic region is mostly disordered and could gain structure in normal conditions within the body.
With increasing temperature, the beta-strands in the cytosolic region increased to 3-4 strands, with proper folding. At very high temperatures, that is, at 410K, some of these changes were reversed, leading to a reduction in the total secondary structure element (SSE). Considerable fluctuations occur along with structural changes.
What are the implications?
Gain of structure, as shown here by the adoption of β-sheets as temperatures rise, is typical of an intrinsically disordered protein region (IDPR) when it interacts with its partner, or under the normal conditions of the body. When the IDPR, in this case, the cytosolic domain, is unstructured, it acts as a MoRF in order to bind transporting vesicles coated with COP1, and thus to allow the spike to localize within the ER.
The researchers quote an earlier report suggesting that the ER localization signal is consistently active in the unfolded protein conformation, but when it is properly folded, it interacts with the M protein, which in turn promotes its incorporation into the virion. More spike molecules are attracted to the surface of the cell by as yet unknown retention signals. These cause syncytia to form, allowing the infection to spread.
With the SARS-CoV-2 also, these researchers have shown that the C-terminal domain, as well as the S2 subunit, interact with the M or membrane protein of the virus through stable intermolecular bonds. Such interactions are likely in the unstructured or disorderly form of the cytosolic domain. The presence of multiple conformations during the period of simulation may allow a number of hypotheses about the function of the spike protein to be explored.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources