The emergence of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants has marked a new phase in the COVID-19 pandemic. Phylogenetic and epidemiological investigations suggest that the SARS-CoV-2 genetic lineage B.1.1.7 may be associated with increased viral transmissibility among humans. It was also classified as a "Variant of Concern" (VOC B.1.1.7) in December 2020. First discovered in Kent, United Kingdom, in September 2020, this new variant has since been found in more than 29 countries across the globe, including the USA.
On January 7, 2021, the team downloaded all B.1.1.7 lineage SARS-CoV-2 genomic sequences available on the GISAID public database - about 8,786 full-length genome sequences from 29 countries. The majority (96.4%, n=8468) of the sequences were from the UK, and about 318 sequences were from other countries, with 13 from North America - 8 from the US and 5 from Canada.
"To estimate the timing of the introduction of B.1.1.7 variants outside the UK, we applied a multistep analytic approach as previously described by our group for HIV."
The researchers combined these B.1.1.7 sequences with a representative set of non-B.1.1.7 sequences (n=3,163) on the basis of sequence homology. The last set of 11,949 sequences was aligned with MAFFT7, and a Maximum Likelihood phylogeny was inferred using IQ-TREE v2.1.28.
The resultant phylogeny showed that all B.1.1.7 samples available group together with high support. Non-UK VOC B.1.1.7 sequences mingled with those from the UK. Since convergent evolution can cause incorrect clustering, the same approach was repeated after excluding variable positions that define the B.1.1.7. lineage and the results were similar.
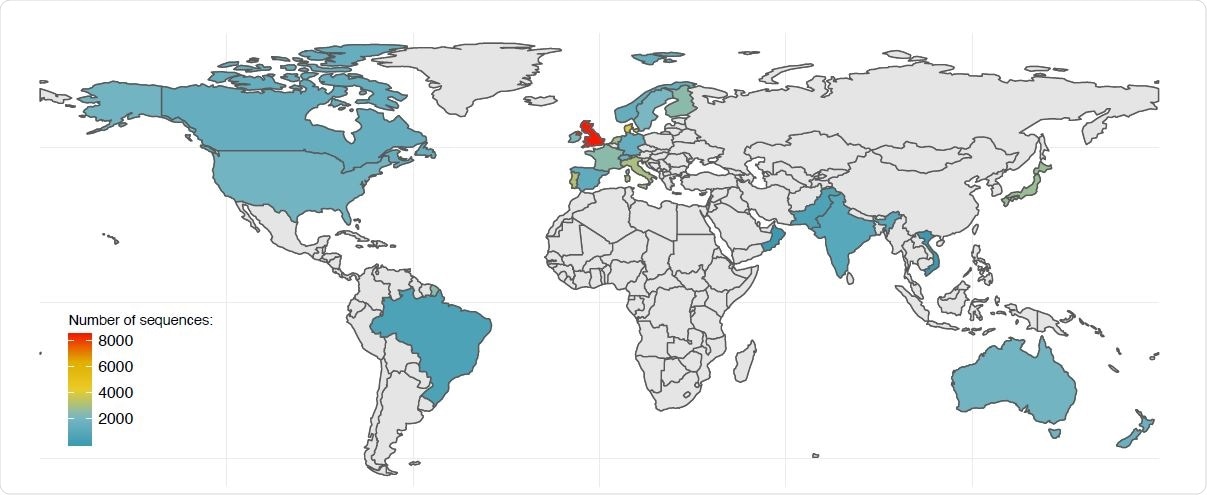
Map of the B.1.1.7 genomic sequences available on GISAID as of January 7, 2020. Countries are colored based on the number of publicly available B.1.1.7 sequences.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Results show that SARS-CoV-2 B.1.1.7 variant first originated in the UK and spread across the globe
The study results did not suggest that the established mutations of VOC B.1.1.7 independently evolved in many different locations. The findings' patterns agree with the view that the new variant first emerged in the UK and spread across the globe.
The researchers believe that the initial and subsequent global spread of the VOC B.1.1.7 variant was facilitated by global connectedness and increased human mobility. The rapid global spread of the new variant VOC B.1.1.7 shows that restrictions currently in place are insufficient to prevent the spread of new and emerging variants of SARS-CoV-2.
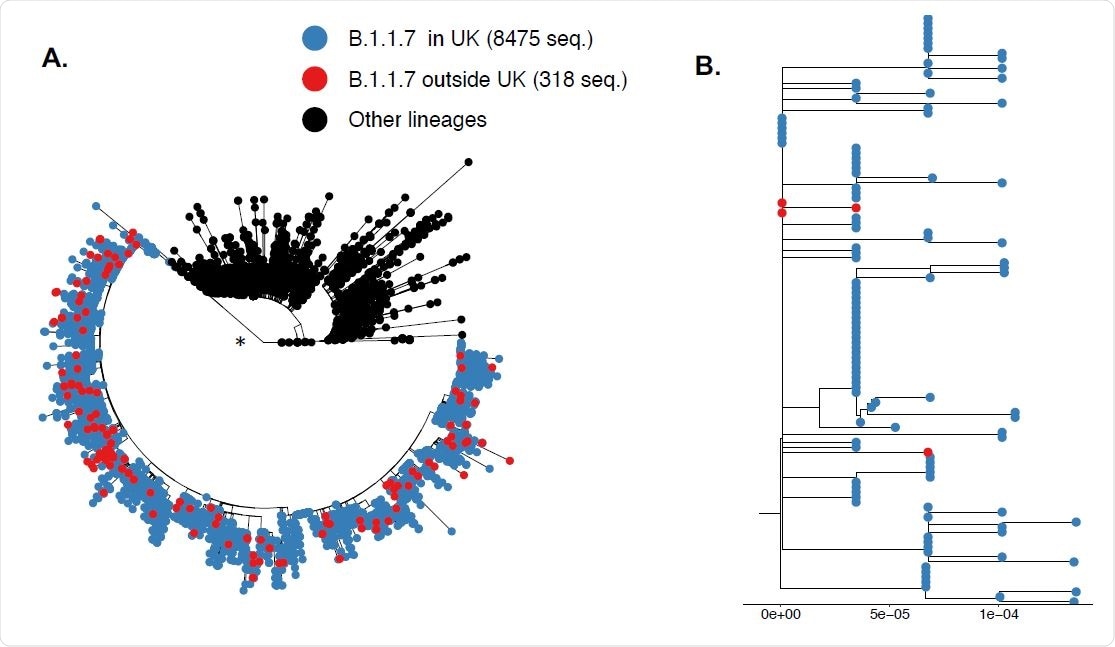
SARS-CoV-2 variant B.1.1.7 arose in the UK and spread globally from there. Tips in the phylogeny are colored according to lineage and country of origin (blue denotes taxa from UK, red denotes taxa from outside UK, and black corresponds to other lineages). (*) The branch leading tot the VOC B.1.1.7 clade has close to perfect suppport (0.99 Shimodaira Hasegawa (SH) branch support9-11). A. All sequences included. B. Illustrative subset of B 1.1.7 taxa from UK (in blue) and other countries (in red). The branch length scale (s/s/y) is indicated at the bottom. Tree display was obtained with the R package “ggtree”13.
According to the authors, similar to viruses such as Ebola, HIV, and HCV, actions to contain the spread of SARS-CoV-2 should be created with a broader perspective beyond the national level. Although local reductions in SARS-CoV-2 infection may be successful in the absence of population immunity, imported infections from other countries can still start new waves of viral transmission, likely aggravated by new and possibly more aggressive phenotypic characteristics of the imported viral strains.
"Similar to Ebola, HCV, and HIV, countermeasures to SARS-CoV-2 spread should be developed with a broader perspective than the national level. Otherwise, without population immunity, successful local reductions in SARS-CoV-2 burden will be counteracted by imported infections that set off new waves of viral spread, possibly exacerbated by novel phenotypic characteristics of the imported strains."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources