Viral infection from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes a dysregulated immune response, including low amounts of interferon (IFN) protein needed to suppress viral spread in the body.
New research led by Roy Parker of the University of Colorado Boulder finds that SARS-CoV-2 prevents the IFN response despite an increased presence of IFN-encoding mRNAs by suppressing transcriptional processes. Inhibition of IFN mRNAs triggers degradation from the activation of RNase L and the SARS-CoV-2 protein Nsp1.
The team writes:
“We observed that both type I and type III IFN-encoding mRNAs predominately localize to their transcriptional sites in a majority of cells. Since this effect is atypical of IFN induction and not a consequence of RNase L activation in response to poly(I:C), we suggest that SARS-CoV-2 inhibits some aspect of RNA processing or an early step of mRNA export, either of which is necessary for efficient release of stable mRNAs from transcription sites.”
The researchers suggest their results could help in COVID-19 with the development of drugs that boost the innate immune response by preventing mRNA decay.
The study “Rapid decay of host basal mRNAs during SARS-CoV-2 infection perturbs host antiviral mRNA biogenesis and export” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
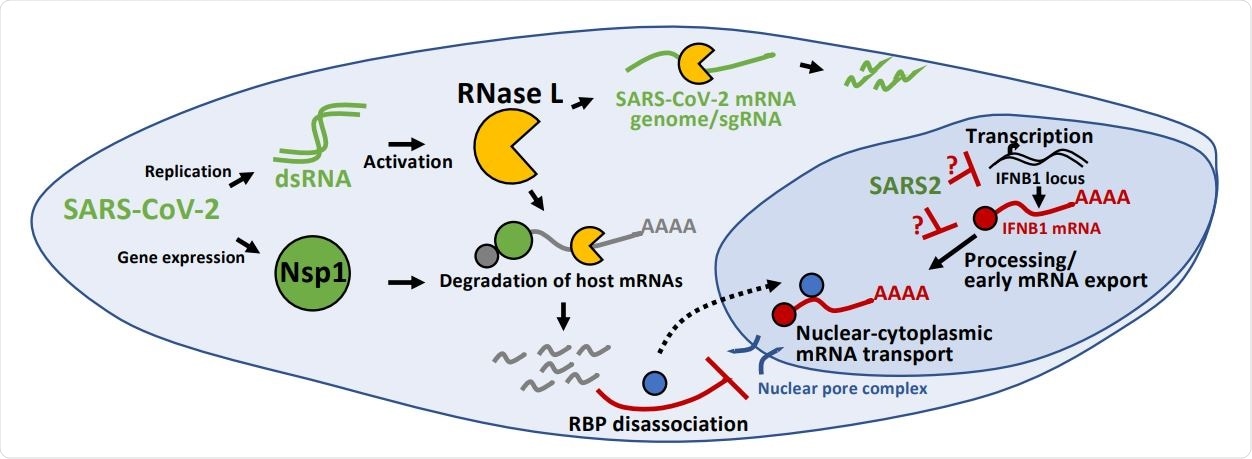
Inhibition of antiviral mRNA biogenesis during SARS-CoV-2 infection. Schematic modeling how antiviral mRNA biogenesis is inhibited during SARS-CoV-2 infection. SARS-CoV-2 replication generates double-stranded RNA (dsRNA), which leads to RNase L activation. RNase L-mediated mRNA decay reduces SARS-CoV-2 full-length mRNA genome and sub-genomic mRNAs. In addition, SARS-CoV-2 expresses the viral Nsp1 protein. Both RNase L activation and Nsp1 expression result in rapid and widespread decay of host basal mRNAs. While RNase L directly cleaves mRNAs, the mechanism of Nsp1-mediated mRNA decay is unclear. The degradation of host mRNAs results in release of RNA-binding proteins (RBPs), and this perturbs late stages of nuclear-cytoplasmic RNA transport. The sequestration of antiviral mRNAs, such as IFNB1 mRNA, in the nucleus prevents their association with ribosomes in the cytoplasm, reducing their translation for protein production. In addition, SARS-CoV-2 inhibits the transcription, an aspect of mRNA processing, or association with early mRNA export factors, and/or rapidly degrades dsRNA-induced antiviral mRNAs, such as IFNB1 mRNA. The result of this is the inability of IFNB1 mRNAs to exit the site of IFNB1 transcription, preventing their transport to the cytoplasm and reducing their translation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Identifying SARS-CoV-2 in host cells
Researchers analyzed viral infection on host cells and how they affect host basal IFN mRNAs. They hypothesized that IFN protein production was being blocked by RNase L-mediated inhibition of mRNA export which activates with the presence of SARS-CoV-2. Using A549 lung carcinoma cell lines with an ACE2-encoding lentivirus, they transduced a wild-type strain with an RNase L knockout.
In response to infection, the RNase L degraded GAPDH mRNA and nuclear retention of IFNB mRNA. This confirmed an innate immune response of A549 cells expressing the ACE2 receptor to double-stranded RNA.
Researchers identified cells infected with SARs-CoV-2 by probing for specific targets, including ORF1a, ORF1b, or N regions of the SARS-CoV-2 genomic mRNA. They then stained the lung cells multiple times after infection and found ORF1a and N RNA after four hours. After eight hours, the virus’s genome was approximately 10-fold higher, with sub-genomic RNA found all over the host cell.
Widespread host mRNA degradation
SARS-CoV-2 decreased GADPH and ACTB mRNA levels eight hours after infection. However, this process could occur independently of RNAase L-mediated mRNA decay. Using RNAase L-knockout cells that removed the enzyme, they continued to find a reduction in GADPH and ACTB mRNA.
RNase L can also be activated by SARS-CoV-2 infection. While it helped to reduce SARS-CoV-2 replication by almost four-fold, it also reduced FL-genome and N-RNA by nearly three-fold.
The activation of the SARS-CoV-2 protein Nsp1 could initiate degradation of host basal mRNA levels, possibly by inhibiting translation. Nsp1, but not Nsp15, decreased ACTB and GADPH mRNAs.
Changes to IFN mRNA during infection
Based on results of SARS-CoV-2 having two mechanisms for basal mRNA degradation — activation of RNase L and Nsp1 — they next looked to see how this affected degradation for IFN during COVID-19 infection.
The team observed the virus triggers RLR-MAVS-IRF3/7 signaling to start transcription of IFN genes. However, about 55% of viral-infected cells lacked IFNB1 expression. Although the researchers note this might be more due to the heterogeneity in the immune response of A549 cells.
They found greater than 82% of induction of IFNB1 in SARS-CoV-2 infected cells, but less than 18% of infected cells had diffuse IFNB1 mRNAs. To determine if RNAse L was causing this, the researchers observed cells with the RNAse L enzyme and found 94% of cells had widespread dissemination of IFNB1 mRNA in the nucleus or cytoplasm. However, less than 6% had IFNB1 transcriptional foci but lacking disseminated IFNB1 mRNA, confirming that RNase L is not the reason behind this.
Instead, the researchers suggest SARS-CoV-2 infection is the cause for blocking the release of IFN mRNAs and retaining them at sites of transcription or sites of nuclear degradation upon leaving their transcription sites.
SARS-CoV-2 also had a second mechanism by blocking nuclear export of IFN mRNAs, regardless of RNase L activation. “Regardless of the nuclease responsible for mRNA destruction, the nuclear retention of IFN mRNAs away from ribosomes would consequently reduce IFN protein production in response to SARS-CoV-2 infection.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources