The worldwide outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the novel pathogen responsible for the coronavirus disease 2019 (COVID-19) pandemic, has stimulated intensive research into potential antiviral drugs and vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Hepatitis C virus vs. SARS-CoV-2
The hepatitis C virus (HCV) is also a plague of humankind, because of its marked propensity to cause chronic hepatitis. However, it is quite different from SARS-CoV-2, having a narrow range of hosts and a chronic course, unlike the short-term disease seen with the latter, and its broad host tropism. Moreover, HCV is a flavivirus, while SARS-CoV-2 is a coronavirus.
Nonetheless, both are alike in having a single strand of genetic material called ribonucleic acid (RNA). In both cases, their replication involves the formation of membranous sacs, called double-membrane vesicles (DMVs), situated in the cytoplasm.
DMV biogenesis
The formation of these DMVs is elicited by viral-host protein interactions, and as these build up in infected cells, they are considered to be replication organelles. They resemble autophagosomes in outward form, but whereas the latter is meant to engulf cellular debris to break them down, viral DMVs serve the purpose of providing a safe place for viral RNA replication.
DMVs are produced from the endoplasmic reticulum (ER) of the host cell, in response to viral non-structural proteins. The current study sought to identify the common substrates utilized by both viruses during the process of replication.
In HCV, the DMVs were found to be composed of acylglycerolphosphate acyltransferase (AGPAT) 1 and 2, both of which were found to accelerate the synthesis of PA
Phosphatidic acid
Phosphatidic acid (PA) is the molecule that forms the basic building block of di- and tri-acylglycerols as well as all glycerophospholipids. It is also a signaling molecule and incorporates new proteins into membranes. With its highly charged head group, it causes membranes to curve.
These PA properties seem to be important for DMV formation, leading the researchers to focus on AGPATs. These enzymes are known to be essential for normal lipid metabolism in the body, and when they are impaired, serious neurological and lung disorders are known to occur.
Of the 11 AGPATs in mammal cells, AGPAT1 and 2 were found to be involved in PA synthesis, using acyl groups from lysophosphatidic acid (LPA). The knockout of AGPAT1 and 2 simultaneously caused poor lipid droplet formation. When only one was knocked out, HCV replication was reduced by around 50-70%, while a double knockout led to 90% lower viral replication.
When subgenomic RNA replication was examined, the replication was still further reduced. These findings showed that the site of AGPAT effect was viral RNA replication. When both these enzymes were expressed at stable levels, replication returned to normal levels. This was not the case if either was inactive.
This type of inhibition was not seen with dengue virus or Zika virus, two other flaviviruses which cause invaginations of the ER rather than DMV formation.
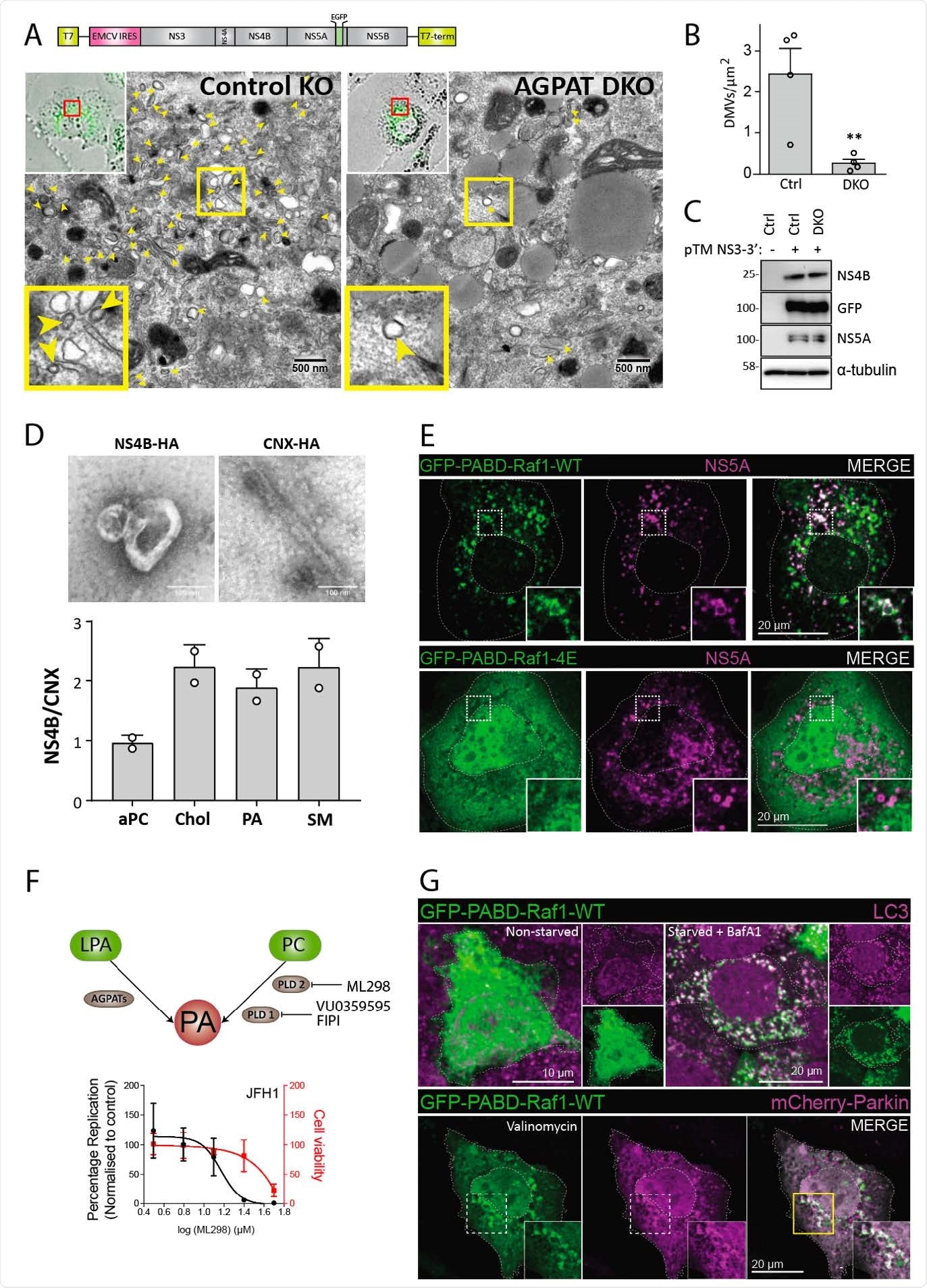
Requirement of AGPATAs for HCV-induced DMV formation and PA. accumulation on HCV-induced DMVs and autophagy-related structures. (A to C) AGPAT1/2 DKO dampens DMV formation induced by HCV. Huh7-derived cells stably expressing the T7 RNA polymerase and containing or not a double knock-out (DKO) of AGPAT1 and 2 were transfected with a HCV replicase-589 encoding plasmid containing a GFP insertion in NS5A (construct pTM NS3-3’/5A-GFP, top panel). Transcripts are generated from the plasmid in the cytoplasm via the T7 promoter and terminator (T7-term) sequence and the HCV NS3 – 5B coding region is translated via the IRES of the encephalomyocarditis virus (EMCV). (A) After 24 h, cells were fixed and subjected to CLEM. Low resolution confocal microscopy images identifying transfected cells are shown on the top left. The area in the red box is shown in the corresponding EM image. Yellow arrow heads indicate DMVs. Insets at the bottom indicate zoomed-in regions. (B) DMVs within whole cell sections were counted and divided by cell area (µm2). Graph shows average and SD from 4 different transfected cells. Cells expressing comparable level of HCV replicase were selected for EM analysis. Significance was calculated by a paired t-test. **, p<0.01. (C) Expression levels of NS4B and NS5A in transfected cells were determined by western blotting. (D) Lipidome analysis of HCV-induced DMVs. Extracts of Huh7 cells containing the subgenomic replicon sg4BHA31R (NS4B-HA) and Huh7 cells stably overexpressing HA-tagged Calnexin (CNX-HA) and control Huh7 cells were prepared as described in supplementary methods and used for HA-affinity purification under native conditions. An aliquot of the sample was analyzed by electron microscopy (top panels) whereas the majority was subjected to lipidome analysis by using mass spectrometry. Values obtained for the NS4B-HA sample were normalized to those obtained for the CNX-HA sample that was set to one. The complete list of analyzed lipids is summarized in data S3. (E) PA accumulation at NS5A containing structures. Huh7-Lunet/T7 cells were transfected with a construct analogous to the one in panel A, but containing a mCherry insertion in lieu of GFP, along with an EGFP-tagged wildtype (WT) or mutant (4E) PA sensor (construct pTM612 EGFP-PABD-Raf1-WT or -4E). Twenty-four hours later, GFP-PABD and NS5A-mCherry were visualized by fluorescence microscopy. White boxes indicate regions magnified in the lower right of each panel. (F) Top panel: Alternate PA biosynthesis pathways via lysophosphatidic acid (LPA) or phosphatidylcholine (PC) catalyzed by AGPATs or PLDs, respectively. Bottom panel: Huh7-Lunet/T7 cells were electroporated with in vitro transcripts of a subgenomic HCV reporter replicon encoding the firefly luciferase. Four hours after transfection, different concentrations of PA synthesis inhibitors were added to the cells and luciferase activities were analyzed at 48 h after electroporation. Graph shows average and SD from 3 independent experiments. Cell viability determined by CellTiter-Glo luminescent assay is indicated with the red line. (G) PA recruitment to autophagy-related structures in selective and non-selective autophagy. Top panel: Huh7-derived cells expressing EGFP623 PABD-Raf-1 were incubated in growth medium (top left panels) or in serum-free medium with 200 nM BafA1 (top right panels) for 3 h. Cells were fixed and stained with a LC3 specific antibody. Bottom panel: For selective autophagy, mCherry-tagged Parkin was co626 expressed with EGFP-PABD-Raf1, followed by incubation with 10 µM Valinomycin to induce mitophagy. Cells were fixed after 3 h, and GFP-PABD and mCherry-Parkin were visualized by fluorescence microscopy. Images in panels E and G are maximum intensity projections.
Inhibition of DMV formation suppresses viral replication
They found that the mechanism of inhibition of RNA replication in HCV was by preventing DMV formation. That is, while high levels of the HCV polyprotein fragment NS3-5B led to abundant DMV formation in control cells, this was prevented to a large extent by double knockout of AGPAT1 and 2.
In addition, the DMVs that were formed were smaller in diameter. Thus, AGPATs are key to the formation of DMVs in vivo. These DMVs are rich in cholesterol and sphingolipids, as well as PA, compared to ER membranes.
Using labeled proteins to detect PA within single cells, it became clear that PA is found in association with NS4B within cells productively infected by HCV. Secondly, they also found that the virus-induced the accumulation of PA at points containing NS5A.
These findings show that PA is recruited to sites of HCV replication, mediated by AGPAT1 and 2.
Alternative pathways of PA inhibition
The effects of inhibition of other enzymes that could play a role in PA generation, such as phospholipase D1 (PLD1) and D2 (PLD2) enzymes, were also examined.
Of three inhibitors used, only one, a PLD2 inhibitor, suppressed replication without causing cytotoxicity. However, this also indicates that PA is required for productive infection with HCV via DMV formation, which supports the replication of viral RNA.
Again, they found that PA is required for autophagosome formation. However, a third pathway involving PA, via diacylglycerol kinase (DAGK), was not found to be instrumental in the formation of these organelles.
Inhibition of SARS-CoV-2-induced DMVs
Since both HCV-induced DMVs and the morphologically similar autophagosomes are both dependent on AGPAT1 and 2, as well as the presence of PA, the study was extended to examine the involvement of these enzymes in the formation of replication organelles of SARS-CoV-2 as well, since these also replicate in DMVs. In this case, DMVs are clumped together with convoluted membranes and zippered ER to form replication organelles.
The findings show that AGPAT1 and 2 are accumulated at DMV biogenetic sites, along with the non-structural proteins nsp3, which are probably the sites of viral RNA replication. This was independent of the drastic reshuffling of ER proteins following SARS-CoV-2 infection, as shown by the intact distribution of other ER proteins.
Similarly, knockout of AGPAT1 and/or 2 caused a marked suppression of SARS-CoV-2 infection in cell culture, indicating reduced viral replication. This was not due to the decrease in the abundance of viral nsp3-5n; neither was there a reduction in the number of DMVs induced by nsp3-4 – unlike that observed with HCV.
Instead, there were alterations in the membrane structure, such as zippered ER and DMVs with an average smaller diameter of 145 nm. Multi-membrane vesicles (MMVs) were found in increased abundance in these cells, indicating that abnormal membrane structures were being formed.
PA is essential for SARS-CoV-2 replication
SARS-CoV-2 replication was less dramatically affected than that observed with HCV, and the absence of AGPAT1 and 2 in cells infected with the former caused the form of the nsp3-4 induced DMVs to change. However, AGPATs still accumulated at the sites where SARS-CoV-2-induced DMV clusters were localized.
This could mean alternative PA biosynthesis pathways were active in allowing DMV formation and SARS-CoV-2 replication. The researchers confirmed this by adding inhibitors of PA synthesis from other lipids, including LPA. This showed a dose-dependent reduction in SARS-CoV-2 replication below cytotoxic concentrations of the drugs.
The best effect was seen with combinations of inhibitors, where viral replication was steeply reduced without cytotoxicity. With all drugs, PA accumulation was reduced at the sites of DMV formation, and these organelles were also markedly smaller. This effect was observed with inhibitors of AGPAT, PLD1, and DAGK.
However, PLD2 inhibition led to the formation of a higher number of MMVs and also increased DMV diameter, as with AGPAT knockout. Thus, this indicates that PA is required for normal DMV formation and RNA replication in SARS-CoV-2 infection.
What are the implications?
Thus SARS-CoV-2 and HCV both form DMVs based on a common lipid molecule, and PA biogenesis also seems to be required for DMVs resembling autophagosomes. This means that cellular DMVs are produced in a similar manner to viral DMVs, and that both are promoted by multiple pathways.
Different mechanisms for the action of PA in viral DMV formation are proposed. It could be that the cone shape of the lipid promotes membrane curvature, predisposing to DMV formation. Secondly, the role played by PA in membrane fission may be related to DMV biogenesis. Finally, it may take part in a counter-transport chain, where it is exchanged for another lipid.
Phosphatidic acid is important for the formation of double membrane vesicles, serving as replication organelles of hepatitis C virus and SARS-CoV-2, and offering a possible host targeting strategy to treat SARS-CoV-2 infection.”
Source

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tabata, K. et al. (2021). Convergent use of phosphatidic acid for Hepatitis C virus and SARS-CoV-2 replication organelle formation. bioRxiv preprint. doi: https://doi.org/10.1101/2021.05.10.443480, https://www.biorxiv.org/content/10.1101/2021.05.10.443480v1
- Peer reviewed and published scientific report.
Tabata, Keisuke, Vibhu Prasad, David Paul, Ji-Young Lee, Minh-Tu Pham, Woan-Ing Twu, Christopher J. Neufeldt, et al. 2021. “Convergent Use of Phosphatidic Acid for Hepatitis c Virus and SARS-CoV-2 Replication Organelle Formation.” Nature Communications 12 (1). https://doi.org/10.1038/s41467-021-27511-1. https://www.nature.com/articles/s41467-021-27511-1.