COVID-19 (coronavirus 2019), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is continuing to cause morbidity and mortality, despite ongoing mitigation strategies and vaccination efforts. Attempts by most countries to combat COVID-19 shave relied on social distancing, lockdowns, limited testing, contact tracing, and vaccine rollouts, all of which are now helping to fight the pandemic.
Aiming to reduce human suffering, economic costs, and restrictions on personal freedom caused by COVID-19 to a minimum, researchers from Germany and Austria proposed an alternative method - efficient population-based genome-based testing.
They demonstrated an affordable, scalable, and highly sensitive virus genome-based testing approach developed as part of the Human Genome Project.
This testing was used to genotype billions of samples over the last decade. Here, they achieved the same sensitivity and specificity, however, in a much more scalable fashion and at much lower costs per sample (~€ 1 per PCR test for very high throughput). The researchers have also provided a link where they share the notes taken in the study, and the results are published on the medRxiv* preprint server prior to peer review.
.jpg)
Schematic of the high throughput genome-based testing pipeline for SARS-CoV-2. (a) Sample tubes (barcoded) are brought to testing centres in 96-well carriers from collection points (e.g. see Figure 2) and prepared for high-throughput testing for SARS-CoV-2 (or other viruses). Samples are heat inactivated before automated transfer in 384-well format (b) and preparation for (c) direct RT-PCR without a requirement for RNA extractions and (d) endpoint measurement showing results of test (positive, negative).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
“The procedure is EU-wide CE (European Conformity) approved in gargle- and smear-based versions and is already being used in Germany to test the employees of Unilever and other companies,” the researchers said in the paper.
Discussing the results of the test, the researchers highlighted the key advantages. Infected individuals can be identified days earlier than with the antigen-based rapid tests currently used. Early detection is a crucial prerequisite to stop the spread of the virus. Another benefit of this test is scalability.
“A single commercially available water bath PCR system with a capacity of 100x 384 PCR plates per run would be able to carry out >600,000 RT-PCR reactions per day, close to three times the entire PCR test capacity currently available in Germany.”
This approach can also be easily used to test for the variants - with a second analysis cycle with a small number of additional tests on the SARS-CoV-2 positive samples.
“In spite of these likely advantages, and an early successful test of the strategy in Vò, an Italian city with 3,300 inhabitants, few governments seem to have seriously considered this as an alternative and, with the exception of Austria (‘Alles gurgelt’), none seem to have supported the establishment of the required infrastructure.”
For proof-of-concept, on a population scale, Vienna, Austria, has approved a program aiming for the early interruption of infection chains, and then contact tracing to prevent the spread of SARS-CoV-2. Individuals self-collected mouthwash samples with the support of a dedicated Web App and verified them (www.lead-horizon.com). The researchers presented an outline (in 5 steps) of how the population-scale testing works in practice, as an example of the decentralized test strategy used in Vienna.
A reliable PCR test result was guaranteed for every inhabitant of the community within 24 hours of the sample collection. In this program, all residents were invited to participate twice a week, as regular testing ensures that people with COVID-19 can be quarantined promptly and chains of infection can be broken as soon as possible. Local drug stores provided validated self-collection kits for mouthwash samples, making PCR tests accessible to all residents within 5 minutes of walking distance. Samples were returned to the lab for transportation, after being packaged in biohazard safe, sealed transport bags and packaging.
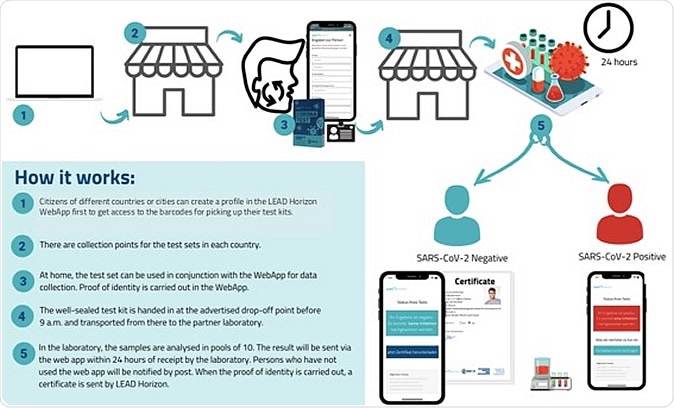
Population-wide testing logistics. An example of the decentralized test strategy used in Vienna.
Vienna provides free sampling devices to all residents, tourists, and commuters. As a result of its success, the program is currently being expanded to more pilot regions of Austria to further evaluate a more comprehensive national coverage. As an alternative, Alacris Theranostics has developed a similar kit that utilizes nasal swabs for high throughput testing.
Using a model based on the established Kermack-McKendrick theory, adjusted to COVID-19, the researchers showed the effect of this population-wide testing approach on the development of the pandemic if it was adopted last autumn. Moreover, assuming certain conditions and removing a few errors, they demonstrated that the model convincingly calculated ‘the effectiveness of mass tests’ - under only moderate contact restrictions like those in Germany in early October 2020.
They suggested that, “Even the rise of the new UK virus variant in early 2021 could have been kept under control by mass testing using highly sensitive PCR based tests, suppressing the R factor from 1.5 before the onset of testing to very low levels for a high degree of participation.”
Importantly, they emphasized this in comparison with modeling the effect of the currently used antigen rapid tests that detect only when an individual becomes positive after the onset of symptoms, which is fairly late in the infectious phase.
From the simulations conducted comparing PCR and antigen-based tests, this study indicated that the population-wide PCR-based tests - even at a fairly low level of participation during the second wave (October 2020), conducted at a mass scale could have resulted in a rapid drop in numbers of infections. It would have then been to manage contact tracing.
To summarize, the researchers have shown in this study that systematic population-wide screens could eliminate COVID-19. However, noting the failure to conduct urgent mass-testing, they recommended that complementing vaccinations, this mass-testing infrastructure could be enabled and tapped to respond to SARS-CoV-2 and also other new similar pathogens.
“The overwhelming majority of this impact, including close to 90% of deaths, was caused by the second and third waves of the pandemic, and could therefore have been potentially avoided by PCR based mass testing.”
Given the enormous impact this approach could have had on the course of the pandemic, this raises the question of the mechanisms governments use to evaluate potential solutions to problems of such enormous importance, should not be more open and science-driven, the researchers write.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Evidence-based pandemic preparedness: an infrastructure for population-scale genome-based based testing for COVID-19, Hans Lehrach, Jon Curtis, Bodo Lange, Lesley A Ogilvie, Richard Gauss, Christoph Steininger, Erhard Scholz, Matthias Kreck, medRxiv 2021.06.10.21258526; doi: https://doi.org/10.1101/2021.06.10.21258526, https://www.medrxiv.org/content/10.1101/2021.06.10.21258526v1
- Peer reviewed and published scientific report.
Lehrach, Hans, Jon Curtis, Bodo Lange, Lesley A. Ogilvie, Richard Gauss, Christoph Steininger, Erhard Scholz, and Matthias Kreck. 2022. “Proposal of a Population Wide Genome-Based Testing for Covid-19.” Scientific Reports 12 (1): 5618. https://doi.org/10.1038/s41598-022-08934-2. https://www.nature.com/articles/s41598-022-08934-2.