Once severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) successfully enters a host cell, the virus adopts host machinery to facilitate replication and proliferation, abrogating host messenger RNA (mRNA) translation in this process. The production of interferon and interferon-stimulated genes within the host cell is often seen to be impaired by this process, limiting the magnitude of the immune response generated.
In a paper recently uploaded to the preprint server bioRxiv* by Schamel et al. (June 25th, 2021) engineered T cells against SARS-CoV-2 are tested against a range of cells in vitro, finding that the treatment could be used to restore cellular immune function and suppress SARS-CoV-2.
Engineered T cells
The group generated T cells reprogrammed to recognize SARS-CoV-2 infected cells using TCR fusion constructs. The TCR is a large multiprotein complex containing antigen-binding and signal transduction subunits. TCR fusion constructs have been well explored in targeted chemotherapy, allowing good preferential delivery of a drug package to the intended cell type, though in this case, the group explored the ability of these cells to counteract viral infection in vitro. The construct bears a complementary ligand to the SARS-CoV-2 spike protein, which is expressed on the surface of host cells to facilitate proliferation, consisting of motifs known to be present on high-affinity antibodies towards the spike protein. This ensures high binding specificity and binding results in a conformational change of the TCR protein machinery that exposes CD3 signaling motifs, which then activates the T cell and initiates cytokine production.
CD4+ and CD8+ T cells were employed in the study, which are both involved in the removal of pathogens by cytotoxic activity. To test the cytotoxic potential of this platform confocal fluorescent imaging was utilized to identify the relative number and position of T-cells, control cells, and target SARS-CoV-2 infected cells. A range of cell types expressing the SARS-CoV-2 spike protein were lysed by the T cells very efficiently, with some lingering effect noted in those expressing the SARS-CoV-1 spike protein due to the remaining similarities between these viral proteins.
Preventing translational shutdown
The production of cytokines IL2, IFNγ, IFNα, and TNFα was seen to be upregulated only by T cells bearing the spike protein-specific TCR machinery, and only in infected cells treated with the proper T cells was luciferase activity maintained, which is usually seen to decline due to SARS-CoV-2 infection. Loss of luciferase signal in the normal course of SARS-CoV-2 is attributed to cell death, reduced proliferation, or suppression of host protein synthesis, and that treatment with the TCR construct T cells maintains and restores luciferase activity in infected cells suggests that these causes are mitigated.
The group found that of the three causes, translational shutdown tends to occur at later time points post-infection, during the viral replication cycle. Loss of luciferase activity at these later time points was most substantially restored by treatment with the T cells, suggesting that it is this process in particular that is prevented by treatment. The group suggests that this is achieved by secretion of a protective factor instead of inducing cell death as observed at some stages of infection.
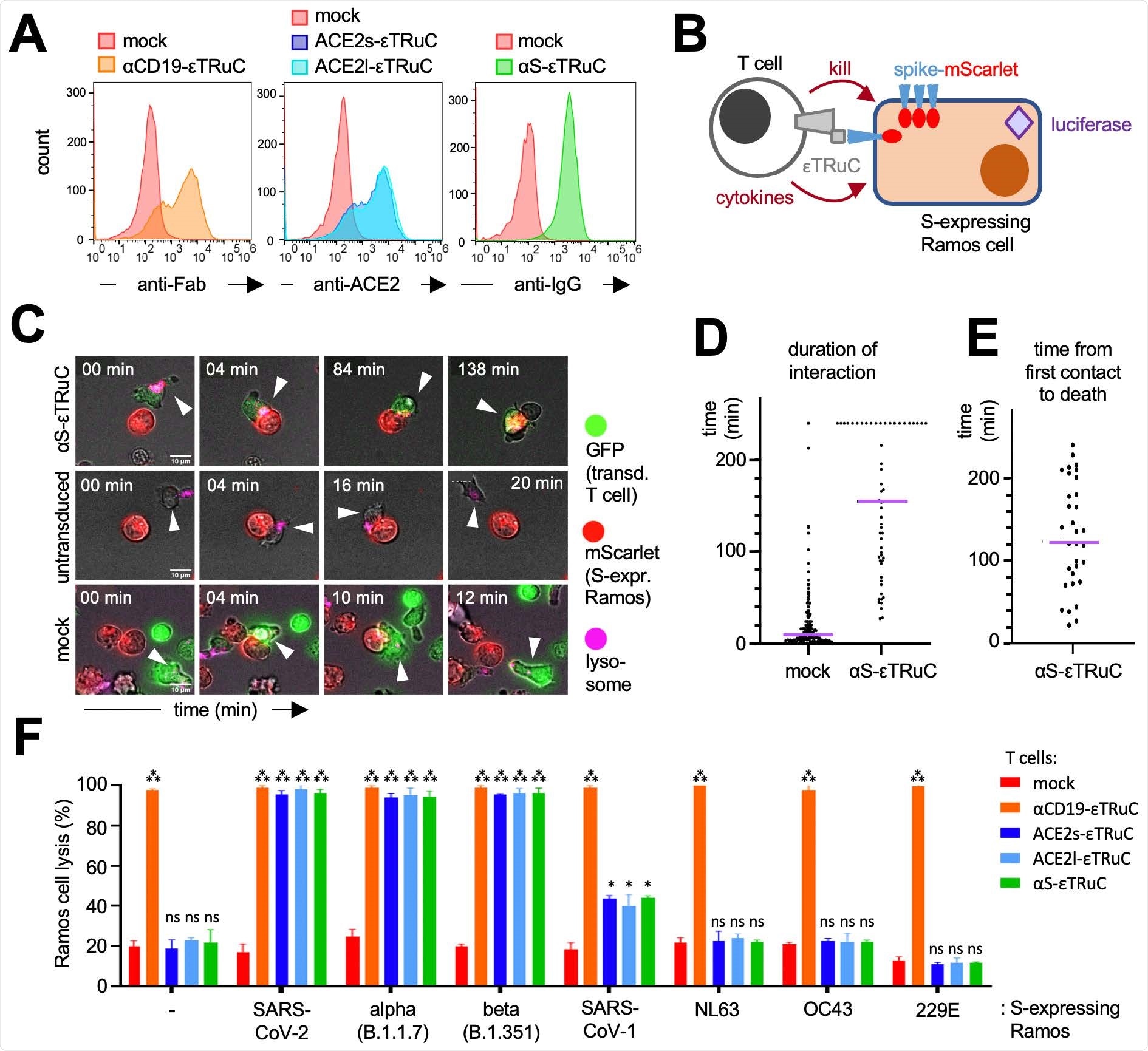
S-specific εTRuC T cells selectively eliminate S-expressing cells. A, Primary human T cells were transduced with the lentiviral vectors encoding for the εTRuCs or the mock vector and expanded with IL-2. Surface expression of the εTRuCs was determined by anti-Fab, anti-ACE2 and anti-IgG staining as indicated. B, S-and luciferaseexpressing Ramos cells are killed by the new εTRuC T cells. C, An αS-εTRuC T cell (green) with the lysosomes stained in pink and an S-mScarlet-expressing Ramos cell (red) were imaged and selected frames of the given times are shown (upper panel). Non-transduced (middle panel) and mock-transduced (green, lower panel) T cells were imaged together with the S-mScarlet-expressing Ramos cells (red). D, Quantification of the duration of interaction between S-mScarlet-expressing Ramos cells and αS-εTRuC or mock T cells from the 4 h videos. E, Quantification of the time it takes for an αS-εTRuC T cell to kill an S-mScarletexpressing Ramos cell (the purple lines in D and E depict the median). F, Ramos cells expressing luciferase, BFP and the different S-proteins were co-cultured with the εTRuC T cells for 24 h at in a 1:1 ratio. Target cell lysis was measured by a loss of luciferase activity in triplicates (the experiment was repeated more than 3 times, n>3).

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Killing events induced by the T cells tended to occur within 30 minutes and 4 hours and was mediated by the quantity of spike protein expressed on the cell surface, and this applied to cells infected with wildtype, B.1.1.7, and B.1.351 SARS-CoV-2 variants. This time is shorter than the average replication time of the virus, 8 hours, though the group observed that the virus was still able to spread in culture, suggested that killing events are insufficient to prevent proliferation. Three days post-infection, cells treated with the T cells had similar levels of luciferase activity as uninfected cells, suggesting that the T cell therapy could be useful as the basis of a COVID-19 therapy focused on restoring cell immunity.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Engineered chimeric T cell receptor fusion construct (TRuC)-expressing T cells prevent translational shutdown in SARS-CoV-2-infected cells, Ira Godbole, Kevin Ciminski, O. Sascha Yousefi, Salma Pathan-Chhatbar, Deniz Saltukoglu, Niklas Vesper, Pavel Salavei, Juliane Strietz, Nicole Gensch, Michael Reth, Martin Schwemmle, Wolfgang W. Schamel, bioRxiv, 2021.06.25.449871; doi: https://doi.org/10.1101/2021.06.25.449871, https://www.biorxiv.org/content/10.1101/2021.06.25.449871v1