Recent advances in next‐generation sequencing (NGS) such as targeted or non-targeted whole-genome sequencing (WGS) and computational analyses that can efficiently process large amounts of data have enabled comprehensive analyses of viral genomes in clinical and research settings. For example, these techniques have been applied to identify causative agents and origins of outbreaks, monitor transmissions, or study the dynamics of epidemics such as Ebola, Zika, and the ongoing severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) pandemic.
The first SARS‐CoV‐2 genomes recovered in early 2020 from 8 patients in Wuhan showed an identity of 99.98% and this high level of shared genomic similarity in viral genomes showed that it is a new virus that has not been circulating for a long time in humans and was possibly a spillover from animals to humans.
Recent studies showed that intra-patient SARS-CoV-2 genetic diversity increased after plasma treatment of patients having high viral loads. Moreover, a high rate of SARS-CoV-2 genome mutation was detected in immunodeficient patients infected with the virus.
Non-targeted whole-genome sequencing is a potent tool that helps comprehensive identifications of constituents of microbes in a sample. Since there is no need to direct the analysis to any kind of identification before genome sequencing, this technique can decrease bias and false-negative results. It also allows the determination of genetic aberrations such as single nucleotide variants, insertions, deletions, and copy number variants, even in the noncoding protein regions of the genome.
Comparing the performance of 4 different random priming amplification methods to recover SARS-CoV-2 RNA
Researchers from the UK recently compared the performance of 4 different random priming amplification methods to recover SARS-CoV-2 RNA. In method 1 (H-P), the reverse transcriptase (RT) step included random hexamers, whereas, in methods 2-4, RT incorporated an octamer primer with a known tag. This study is published on the bioRxiv* preprint server.
In methods 1 and 2, (K-P) sequencing was performed on material derived from the RT-PCR step, while in methods 3 (SISPA) and 4 (S-P), additional amplification was performed before sequencing. They also presented the S-P technique that might be applicable for Oxford Nanopore real-time sequencing as a non-targeted primer-based protocol is not available yet.
“Presented in this study, the SISPA protocol allowed for whole-genome assembly of both viruses using only one primer in a sequence-independent reaction.”
SISPA method is the most efficient method of the 4 methods for whole-genome sequencing of SARS-CoV-2
The results showed that the SISPA method described by the authors in this study was the most efficient method for non-targeted/random priming whole-genome sequencing of SARS-CoV`-2. This method allowed for whole-genome assembly of SARS-CoV-2 and influenza A(H1N1) pdm09 in mixed samples. They determined the detection limit - 103 pfu/ml (Ct, 22.4) for whole-genome assembly - and the limit of characterization of SARS-CoV-2 - 101 pfu/ml (Ct, 30) for metagenomics detection. The whole-genome sequences obtained using the SISPA (or S-P) method in this study were free of primer bias and allowed for polymorphism analysis.
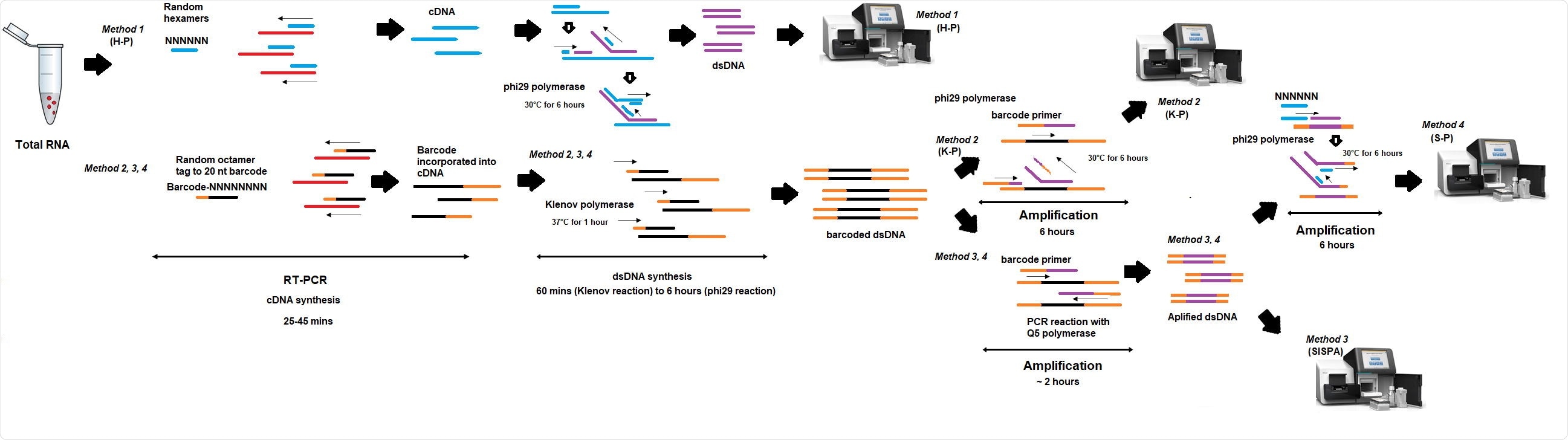
Schema of random amplification methods for whole genome assembly of SARS613 Cov-2 virus genome. Method 1 (H-P) is based on the RT-PCR step with random hexamers primer (6Ns) followed by phi29 polymerase isothermal amplification in the presence of 6Ns primer and then library preparation for Illumina sequencing. Method 2 (K-P), random octamer tagged with 20 nucleotide known tag sequence (5`- GACCATCTAGCGACCTCCACNNNNNNNN-3`) (K-8N) was used for RT-PCR step, followed by phi29 polymerase isothermal amplification in the presence of tagged primer K-8N and then library preparation and Illumina sequencing. Method 3, Sequence-Independent, Single620 Primer Amplification (SISPA) technique, followed by library preparation and Illumina sequencing. Method 4 (S-P), following SISPA amplification (Method 3), phi29 polymerase isothermal amplification in the presence of random hexamers (6Ns) was applied and then used for Illumina sequencing

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The SISPA method is mainly used to obtain genome sequences of RNA viruses or investigating complex clinical samples with mixed infections in a single reaction since no sequence information is required.
According to the authors, this method can be used to monitor genomic changes and evolution of the virus, for fast and simple metagenomics detection, and to assess the pathogen profile of the sample that might help identify other factors that might be involved in the coronavirus disease (COVID-19) infection.
“Previous studies have shown active genetic recombination events in SARS-CoV-2 genomes which may reduce the accuracy of conventional qRT-PCR detection, and thus the primers should be precisely chosen to address these challenges.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A random priming amplification method for whole genome sequencing of SARS-CoV-2 and H1N1 influenza A virus Klaudia Chrzastek, Chandana Tennakoon, Dagmara Bialy, Graham Freimanis, John Flannery, Holly Shelton bioRxiv 2021.06.25.449750; doi: https://doi.org/10.1101/2021.06.25.449750, https://www.biorxiv.org/content/10.1101/2021.06.25.449750v1
- Peer reviewed and published scientific report.
Chrzastek, Klaudia, Chandana Tennakoon, Dagmara Bialy, Graham Freimanis, John Flannery, and Holly Shelton. 2022. “A Random Priming Amplification Method for Whole Genome Sequencing of SARS-CoV-2 Virus.” BMC Genomics 23 (1). https://doi.org/10.1186/s12864-022-08563-z. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08563-z.