Even as emerging variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge the potential to achieve herd immunity by vaccination, a considerable amount of research has been published on promising new molecules that have the potential to be effective against this virus.
 Study: Pharmacological Perturbation of Intracellular Dynamics As A Sars-Cov-2 Antiviral Strategy. Image Credit: Sisacorn / Shutterstock.com
Study: Pharmacological Perturbation of Intracellular Dynamics As A Sars-Cov-2 Antiviral Strategy. Image Credit: Sisacorn / Shutterstock.com

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
Since SARS-CoV-2 was first identified as a novel beta coronavirus, many scientists have focused on finding effective pharmacological interventions to counter its rapid and extensive spread. Several SARS-CoV-2 variants of concern (VoCS) have emerged that show higher transmissibility and/or resistance to neutralization by the antibodies elicited by either the earlier strains of SARS-CoV-2 or vaccination. The emergence of SARS-CoV-2 VoCs has thus posed a challenge in global efforts to minimize morbidity and mortality caused by SARS-CoV-2 infection.
Antivirals such as remdesivir, which is a nucleoside analog that inhibits SARS-CoV-2 in vitro by promoting the formation of a false nucleoside chain, failed to prove highly effective against this virus. The limited efficacy of these compounds has been associated with both their significant side effects and the need to administer them intravenously.
Other direct-acting agents (DAA) that inhibit the viral polymerase enzyme have proven to be unreliable for long-term control. This is because ribonucleic acid (RNA) viruses like SARS-CoV-2 typically change their mode of binding via regular mutations, thus allowing the virus to escape such inhibition. This has raised the need for host-targeting agents (HTA) as well.
HTAs target the host cells that foster viral replication and propagation. Therefore, this is the preferred approach when combined with vaccination and DAA.
The advantages of having a wide variety of antivirals are that repurposed drugs may not always act as projected, or may need to be optimized for more efficient antiviral activity. Alternatively, the side effects of certain antivirals may be unacceptable at the population level. Thirdly, they may be easily escaped by mutations.
Finally, using a cocktail of drugs is always useful in maximizing drug efficiency.
SARS-CoV-2 primarily uses the host cell angiotensin-converting enzyme 2 (ACE2) receptor to gain entry into the body. However, this virus also sometimes requires the transmembrane serine protease TMPRSS2 for cell entry, unless the cells show endosomal acidification. Either way, SARS-CoV-2 enters the cell via a clathrin-mediated mechanism that ultimately results in the fusion of the virus with the cell membrane.
Subsequently, the viral RNA genome, protected by the nucleoprotein, enters the ER for the translation of viral proteins and viral replication.
Study findings
The current study focused on a molecule called RG10, which is a sulfamoyl-phenyl derived compound with antiviral activity equivalent to that of remdesivir. This novel small molecule was not toxic to the cell, but had a half-maximal effective concentration (EC50) of about 1.5 micromolar (μM).
RG10 showed excellent antiviral activity against SARS-CoV-2 in vitro, almost abolishing infection of cells that were previously exposed to this virus. Further, RG10 was found to show inhibitory activity only when added after viral entry, thereby indicating that its mechanism of action is by suppressing viral replication, rather than viral entry.
The scientists observed that the fluorescent-labeled RG10 molecule was clustered in the ER, along with the virus replication complexes, though both were in separate regions. This means that RG10 is likely an HTA.
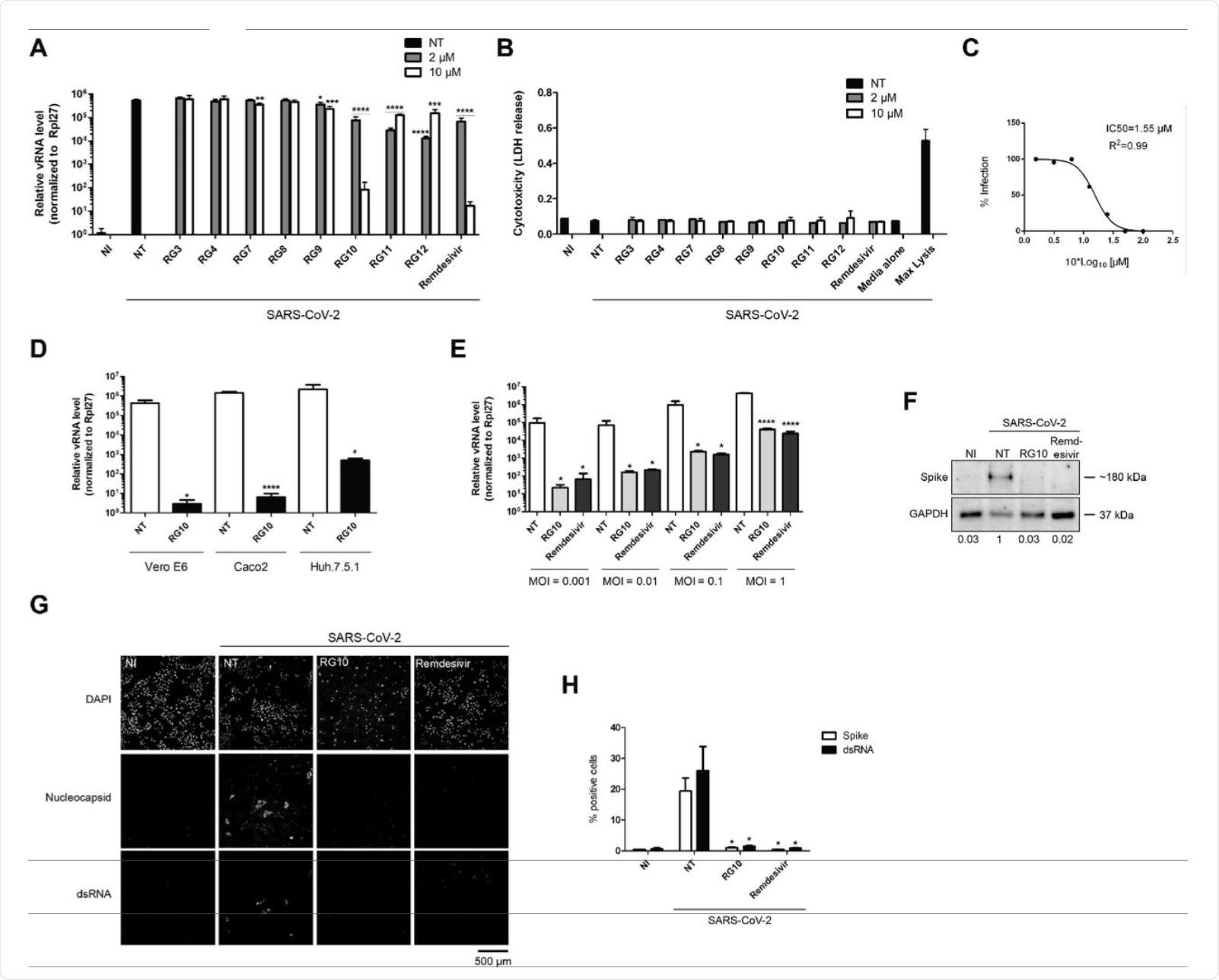 (A-B) Vero E6 cells were treated with the indicated RG derivatives or Remdesivir at indicated concentrations and subsequently infected with SARS-CoV-2 at MOI 0.01 for 48 h in the presence of the compounds. (A) The graph shows relative SARS-CoV-2 RNA (vRNA) levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test ** p value < 0.01; *** p value < 0.005; **** p value < 0.001. (B)Cytotoxicity measured by LDH release assay showed no significant differences between the compounds. (C) Vero E6 cells were treated with RG10 at various concentrations and subsequently infected and processed as in A. The IC50 was 1.55 µM +/- 0.11 as measured from three independent experiments. (D) Vero E6 (monkey kidney cells), Caco2 (human intestinal cells) or Huh7.5.1 cells (human liver cells), were treated with 10 µM RG10 and subsequently infected with CoV2 at MOI 0.01 for 48 h in the presence of the compounds. The graph shows relative CoV2 RNA (vRNA) levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test ** p value < 0.01; *** p value < 0.005; **** p value < 0.001. (E) Huh7.5.1 cells were treated with 10 µM RG10 or 10 µM Remdesivir and subsequently infected with CoV2 at indicated MOI for 48 h. The graph shows relative vRNA levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test * p value < 0.05 and **** p value < 0.001. (F) Huh7.5.1 cells were treated with 10 µM RG10 or 10 µM Remdesivir and subsequently infected with SARS-CoV-2 at MOI 0.5 for 48 h. Cells were then lysed 48 h post infection. The lysates were processed for western blotting using an anti SARS-CoV-2 Spike antibody and an anti-GAPDH antibody as loading control. Both immunostainings were revealed using secondary antibodies coupled to HRP. (G) Cells were infected as in F then fixed and stained using an anti SARS-CoV-2 Nucleocapsid antibody and a J2 antibody recognizing double stranded RNA (dsRNA) representative of viral replication complexes. The micrographs show the individual channels that have been acquired using a confocal microscope and processed using ImageJ. (H) Samples were treated as in F and cells were detached and fixed 48 h post infection. Samples were stained and analyzed by flow cytometry. Student’s t-test * p value < 0.05.
(A-B) Vero E6 cells were treated with the indicated RG derivatives or Remdesivir at indicated concentrations and subsequently infected with SARS-CoV-2 at MOI 0.01 for 48 h in the presence of the compounds. (A) The graph shows relative SARS-CoV-2 RNA (vRNA) levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test ** p value < 0.01; *** p value < 0.005; **** p value < 0.001. (B)Cytotoxicity measured by LDH release assay showed no significant differences between the compounds. (C) Vero E6 cells were treated with RG10 at various concentrations and subsequently infected and processed as in A. The IC50 was 1.55 µM +/- 0.11 as measured from three independent experiments. (D) Vero E6 (monkey kidney cells), Caco2 (human intestinal cells) or Huh7.5.1 cells (human liver cells), were treated with 10 µM RG10 and subsequently infected with CoV2 at MOI 0.01 for 48 h in the presence of the compounds. The graph shows relative CoV2 RNA (vRNA) levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test ** p value < 0.01; *** p value < 0.005; **** p value < 0.001. (E) Huh7.5.1 cells were treated with 10 µM RG10 or 10 µM Remdesivir and subsequently infected with CoV2 at indicated MOI for 48 h. The graph shows relative vRNA levels measured by RT-qPCR and normalized by Rpl27 RNA expression. The bars are means +/- SD from duplicates representative of two independent experiments. Student’s t-test * p value < 0.05 and **** p value < 0.001. (F) Huh7.5.1 cells were treated with 10 µM RG10 or 10 µM Remdesivir and subsequently infected with SARS-CoV-2 at MOI 0.5 for 48 h. Cells were then lysed 48 h post infection. The lysates were processed for western blotting using an anti SARS-CoV-2 Spike antibody and an anti-GAPDH antibody as loading control. Both immunostainings were revealed using secondary antibodies coupled to HRP. (G) Cells were infected as in F then fixed and stained using an anti SARS-CoV-2 Nucleocapsid antibody and a J2 antibody recognizing double stranded RNA (dsRNA) representative of viral replication complexes. The micrographs show the individual channels that have been acquired using a confocal microscope and processed using ImageJ. (H) Samples were treated as in F and cells were detached and fixed 48 h post infection. Samples were stained and analyzed by flow cytometry. Student’s t-test * p value < 0.05.
The antiviral activity of RG10 was not completely explained by the induction of stress on the ER. Instead, this compound slowed down the movement of the virus to the ER for replication and translation. That is, the intracellular dynamics of the infected cell are perturbed by RG10, preventing its smooth transport to the ER.
RG10 did not affect Zika virus infection, which is unlike the molecule tunicamycin, which induces ER stress and affected both SARS-CoV-2 and RG10 equally. RG10 did show activity against the human coronavirus 229E, though with lower potency.
RG10 also showed antiviral efficacy ex vivo in human airway epithelial cells, as demonstrated by its ability to reduce viral infection and viral protein translation. In this model, ER stress was induced by RG10, and the cytoskeletal actin network appeared to show rearrangement at the apical side of the cell.
The RG10 molecule’s stability is low, accounting for its low antiviral potency, which led to the development of a derivative called RG10b. This stable molecule showed an EC50 of less than 1 μM, with a half-life more than three-fold higher compared to the parent molecule.
The potency of RG10b was also higher than that of RG10. To this end, ex vivo challenge with SARS-CoV-2 followed by treatment with RG10b demonstrated a potency equivalent to that of remdesivir.
Implications
The findings of this study demonstrate a potentially useful broad-spectrum inhibitor of SARS-CoV-2 movement to the ER. As a result, RG10b appears to inhibit both viral replication and viral protein translation, all the while maintaining high stability in vivo without causing any toxic effects to arise in the culture cells.
The exact target of RG10 remains unknown; however, it perturbs SARS-CoV-2 and ER dynamics, thus disrupting intracellular trafficking. The actin cytoskeleton also appears to become disorderly to some extent in primary HAE cells after exposure to RG10. However, unlike other actin disruptors like Latrunculin, jasplakinolide, and cytocalasin D, RG10b does not show significant cytotoxicity.
The change in ER morphology following RG10 exposure was another finding. This change in morphology may contribute to its antiviral effects.
The ER needs to be constantly reshaped in order to fulfill the range of functions it sustains, thus allowing the ER membranes to be a prime target for pathogens. Therefore, a dynamic shift in ER morphology could be an antiviral strategy; however, further research is needed to confirm this mechanism.
Atlastins are a class of compounds that also affect the ER. In fact, Zika virus replication is reduced by abnormalities in the ER shape mediated by atlastins. The role played by atlastins in SARS-CoV-2 infection, if any, has not been studied so far.
The researchers of the current study also added to the array of new tools to study how SARS-CoV-2 affects the infected cell. Their use of bright SARS-CoV-2 particles that remain as infectious as the wild-type virus could be imitated in many different assays to understand the different ways in which this virus enters the host cell.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.