A landmark study uncovers how a specific lung gene, FOXP4, raises the risk of persistent symptoms after COVID-19, providing fresh insight into why some people are more susceptible to long COVID than others.
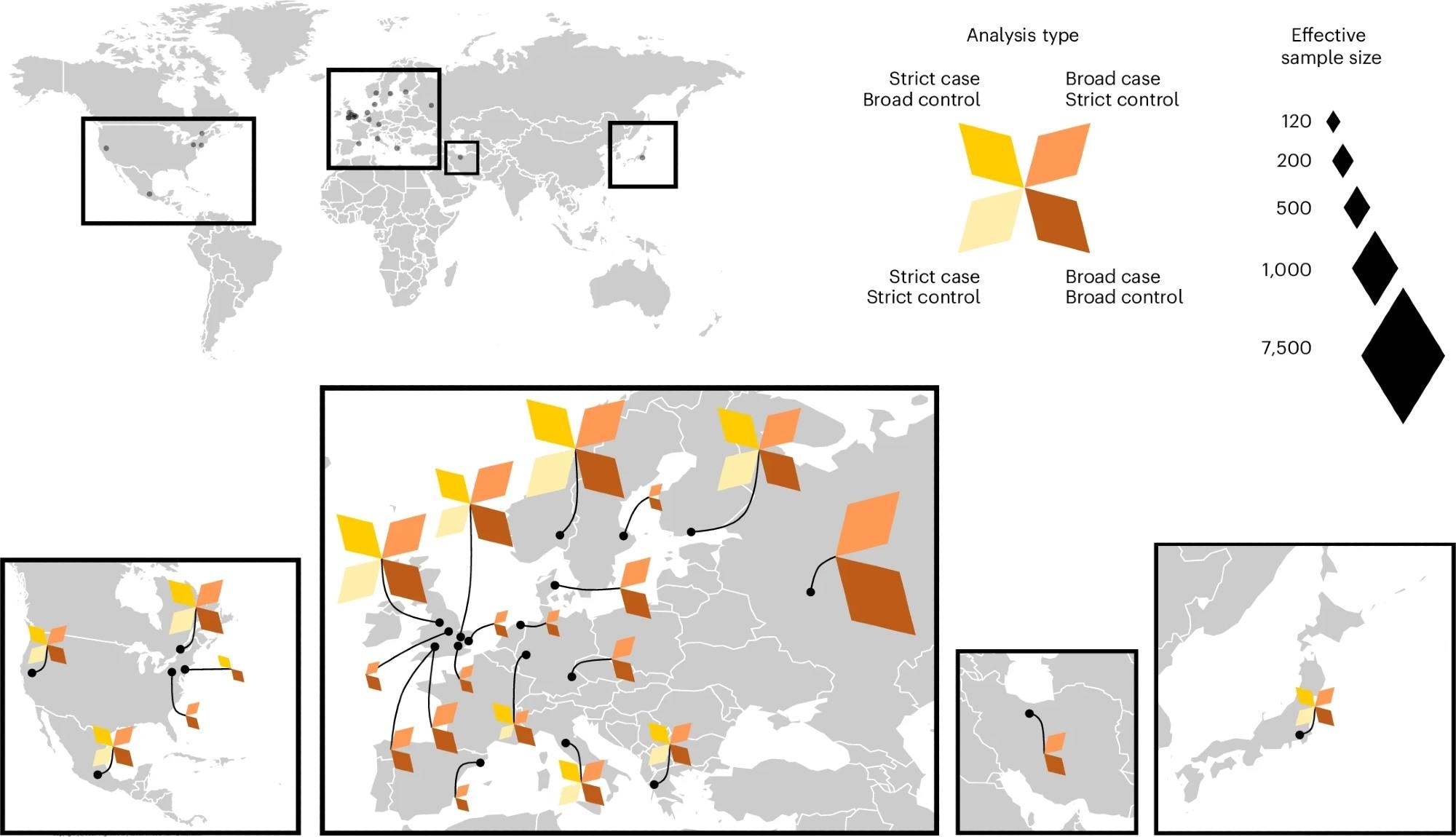 The 24 studies contributing to the Long COVID HGI data freeze 4 served as the discovery cohorts for the GWAS meta-analyses. Each color represents a meta-analysis with specific case and control definitions. Strict case definition, long COVID after test-verified SARS-CoV-2 infection; broad case definition, long COVID after any SARS-CoV-2 infection; strict control definition, individuals that had SARS-CoV-2 but did not develop long COVID; broad control definition, population control, that is, all individuals in each study that did not meet the long COVID criteria. Effective sample sizes are shown as the size of each diamond shape, and locations of sample collection in (from left to right) North America, Europe, Middle East and Asia.
The 24 studies contributing to the Long COVID HGI data freeze 4 served as the discovery cohorts for the GWAS meta-analyses. Each color represents a meta-analysis with specific case and control definitions. Strict case definition, long COVID after test-verified SARS-CoV-2 infection; broad case definition, long COVID after any SARS-CoV-2 infection; strict control definition, individuals that had SARS-CoV-2 but did not develop long COVID; broad control definition, population control, that is, all individuals in each study that did not meet the long COVID criteria. Effective sample sizes are shown as the size of each diamond shape, and locations of sample collection in (from left to right) North America, Europe, Middle East and Asia.
In a recent study in the journal Nature Genetics, researchers conducted a genome-wide association study (GWAS) to elucidate the biological mechanisms governing long COVID development. The study meta-analyzed data from 33 independent long COVID GWAS cohorts comprising a total of 15,950 long COVID cases and 1,892,830 controls across 19 countries.
Study findings highlight FOXP4 gene variants for their statistically significant risk association with long COVID, an association found to be independent of FOXP4's previously identified link to severe COVID-19, thus underscoring the role of lung pathophysiology in developing the condition. Mendelian randomization analyses further identified COVID-19 infection severity as a risk factor in long COVID incidence. Genetically predicted smoking was also explored, but its association was only nominally significant and did not survive correction for multiple comparisons. This suggests the condition is heterogeneous and governed by individual-specific interplays between genetics and environmental exposures, with the study also noting generally low heritability estimates for long COVID (ranging from 0.97% to 12.36% depending on case/control definitions), underscoring the substantial role of non-genetic factors.
Background
The coronavirus disease of 2019 (COVID-19) outbreak remains the worst pandemic in recent human history, causing unprecedented mortality, infrastructure losses, and socioeconomic disruptions. While government-enforcement social distancing measures and the prompt development and distribution of anti-COVID-19 vaccines helped curb disease spread and manage the pandemic, a large proportion (10-70%) of survivors reported persistent COVID-19 symptoms lasting for months or even years, severely degrading their quality of life (QoL).
This new condition, termed ‘post-acute sequelae of COVID-19 (PASC)’ or just ‘long COVID,’ is characterized by symptoms such as fatigue, dysautonomia, pulmonary dysfunction, cognitive disturbances, and others that present within three months following COVID-19 recovery and persist for two months or more (World Health Organization [WHO]).
Several studies have attempted to assess the risk factors associated with the condition, revealing that disease severity increases long COVID risk. Unfortunately, the biological (genetic) underpinnings of long COVID and the mechanisms governing its development remain poorly understood. Consequently, the International COVID-19 Host Genetics Initiative (COVID-19 HGI) was established to investigate the relationship between genetics, disease susceptibility, and severity.
About the study
The present study aims to leverage genome-wide association study (GWAS) to unravel the biological mechanisms (genetic variants) governing long COVID risk using meta-analytic approaches. It also evaluates potential overlap between long COVID-associated genetic variants and those linked to other conditions (e.g., autoimmune diseases, psychiatric disorders) to understand shared heritable architectures.
Study data was aggregated from COVID-19 HGI studies (n = 33) across 19 countries and comprised a total of 15,950 long COVID cases and 1,892,830 controls. Of these, 24 studies contributed to initial discovery analyses, with the primary FOXP4 finding emerging from a meta-analysis involving 3,018 long COVID cases (defined by test-verified SARS-CoV-2 infection) and 994,582 population controls. The largest meta-analysis in the discovery phase included 6,450 long COVID cases (broader definition) and 1,093,955 population controls. The remaining nine studies (9,500 long COVID cases and 798,835 population controls) were used for replication analyses of identified genetic variants.
WHO guidelines were used to classify participants into cases or controls. Controls were further subclassified into ‘population controls’ (all genetic ancestry-matched participants without long COVID) and ‘case controls’ (COVID-19 survivors without long COVID).
Since each included study independently performed sample collection, genotyping, quality control, and outcome (association) analysis, GWAS summary statistics (age, sex, genetic principal components, etc.) were used for this study’s meta-analyses. A modified version of the meta-analysis pipeline described in the COVID-19 HGI’s flagship paper (Nature, 2021) was used to investigate genetic correlations (Linkage Disequilibrium Score Regression v1.0.1).
Causal effects were established using Mendelian randomization (MR) analyses of pooled fixed-effects Inverse Variance Weighted (IVW) data. Significant loci identified herein were annotated using publicly available databases. Finally, pathway enrichment analyses (for biological process discovery), replication analyses (for outcome reliability), and polygenic risk scores (for cumulative long COVID risk assessment) were carried out.
Study findings
Initial discovery meta-analyses identified genetic variants of the FOXP4 gene as significant risk factors for long COVID, an association found to be independent of FOXP4's known link to severe COVID-19, with the C allele at rs9367106 highlighted as the lead variant (odds ratio [OR] = 1.63). The frequency of this risk allele (rs9367106-C) was found to vary considerably across different genetic ancestries, ranging from 1.6% in non-Finnish Europeans to 36% in East Asians, impacting statistical power in different populations.
Replication analyses confirmed these findings for the lead variant rs9367106 (e.g., OR = 1.13 in one independent sample, and OR = 1.21 in the MVP cohort, where rs12660421 also replicated with a similar effect size). Further fine-mapping efforts pointed to rs9381074 as a likely causal variant within the FOXP4 locus, showing relevance across several distinct ancestries, suggesting its functional relevance in condition pathophysiology. Notably, homozygosity for the FOXP4 risk allele was associated with a particularly increased risk for long COVID (OR = 5.64 in one analysis).
FOXP4 is a transcription factor predominantly expressed in lungs and immune cells and has previously been associated with severe COVID-19 infections and lung cancers. Its identification in the present study emphasizes the strong association between pulmonary pathophysiology and long COVID. Observational data showed that long COVID patients had significantly higher FOXP4 gene expression levels in blood than controls, and genetic analyses suggested that variants influencing FOXP4 expression are causally linked to long COVID.
MR analyses further identified COVID-19 infection severity as a causal risk factor in long COVID development. Genetically predicted smoking was also explored as a potential risk factor and showed a nominal association, though this did not survive correction for multiple comparisons. This suggests that at least some of long COVID’s risk factors are potentially modifiable and may be clinically addressed in the future. Vaccination was generally supported by the study as having a protective effect against long COVID, consistent with earlier epidemiological observations. The paper noted that in a specific sub-analysis, the association of the FOXP4 risk allele with long COVID was not significant post-vaccination, but this analysis involved a small sample size of only 40 individuals diagnosed with long COVID following immunization, limiting firm conclusions for that specific genetic variant in the vaccinated group.
Furthermore, the study observed a possible stronger risk associated with FOXP4 risk alleles before widespread vaccination and with earlier viral strains, such as the wild-type and Alpha variants.
Conclusions
This study identified strong genomic evidence linking pulmonary pathophysiology to long COVID, underscoring the importance of FOXP4 (a gene also linked to lung cancer and severe COVID-19) and pulmonary pathophysiology in condition development. It also found causal associations between COVID-19 severity and long COVID, while also highlighting nominally significant associations for potentially modifiable risk factors like smoking, highlighting potential research avenues and future interventions.
The findings also point to the complexity of long COVID, with varying genetic risk allele frequencies across ancestries, a potential increased risk with homozygosity at the FOXP4 locus, possible differential risk based on viral strain and vaccination timing, and overall low heritability, suggesting a significant interplay with environmental factors.
Journal reference:
- Lammi, V., Nakanishi, T., Jones, S. E., Andrews, S. J., Karjalainen, J., Cortés, B., E., H., E., B., Broberg, M., Haapaniemi, H. H., Kanai, M., Pirinen, M., Schmidt, A., Mitchell, R. E., Mousas, A., Mangino, M., Cirulli, E. T., Vaudel, M., Kwong, A. S., . . . Ollila, H. M. (2025). Genome-wide association study of long COVID. Nature Genetics, 1-16. DOI: 0.1038/s41588-025-02100-w, https://www.nature.com/articles/s41588-025-02100-w