
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Although most coronavirus disease 2019 (COVID-19)-infected people completely recover from the disease, there isn't much data regarding the long-lasting or delayed neurological consequences of SARS-CoV-2 infection. Loss of smell and other neurological deficits have been reported during acute SARS-CoV-2 infection. Additionally, several reports have implied the potential risk of Parkinson's disease and other neurodegenerative disorders linked with SARS-CoV-2.
A probable mechanism of Parkinson's disease following COVID-19 is the SARS-CoV-2-mediated amyloid generation as aggregates of aS. In vitro reports suggest that SARS-CoV-2 accentuated aS amyloid formation by interaction with amyloidogenic regions on the nucleocapsid (N), envelope (E), and spike (S) proteins.
A mechanism similar to Alzheimer's disease has been hypothesized for the enhanced amyloid formation leading to Parkinson's disease in COVID-19, which involved the formation of amyloid fibrils as an immune response to infection leading to entrapment and neutralization of the pathogen. However, in-depth information about the exposure to SARS-CoV-2, the appearance of fibrils, and resulting disease symptoms are not available.
About the study
In the present study, the researchers examined how the interaction of the SARS-CoV-2 residual fragment named SK9 located on the C terminal of the E protein impacts the conformational ensemble of aS monomers and the stability of two resolved fibril polymorphs called the rod and the twister structures.
A helix-rich model of the aS monomer structure was resolved employing nuclear magnetic resonance (NMR) solution in the micellar environment and was stored in the Protein Data Bank (PDB) under the identification code:1XQ8. The team evaluated the binding of the SK9 with aS and whether it alters the aS's conformational ensemble by employing a complementary set of molecular dynamic simulations. For assessing the stability of the aS fibrils rod and twister polymorphs, decamers made of five layers and two protofilaments were generated using cryogenic electron microscopy (cryo-EM) structures.
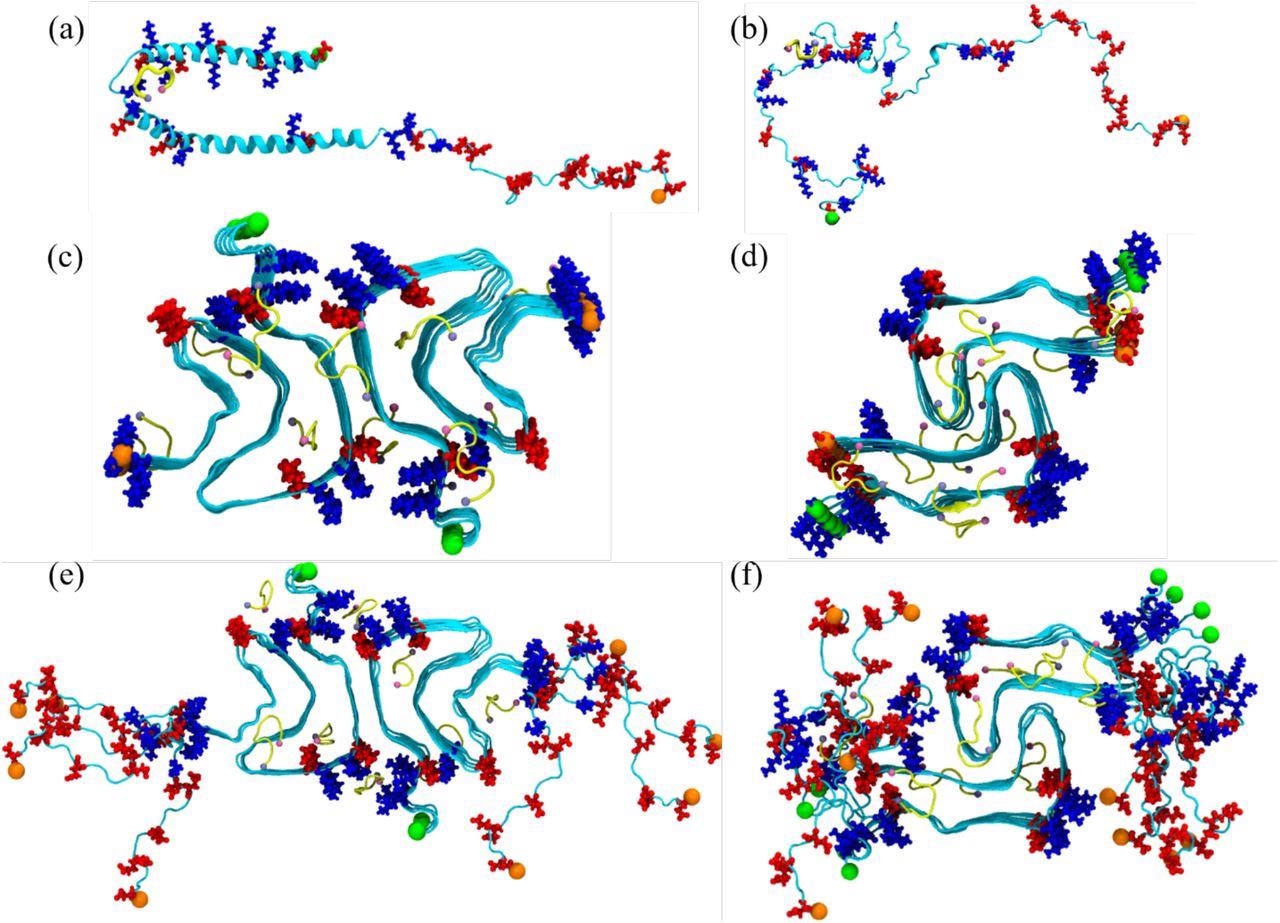
Initial conformation of the α-synuclein monomer (a) as resolved by solution NMR (PDB ID: 1XQ8), and (b) after heating at 500 K to obtain a randomized stretched conformation. Initial conformation for the fibril as derived by cryo-EM structures are shown in (c) for the rod (PDB ID: 6CU7) and in (d) for the twister (PDB ID: 6CU8) polymorph. In (e) and (f) are the corresponding structures shown for the fibrils where the individual chains are extended to residues 38-120. Acidic residues are colored in red and basic ones in blue, while the SK9-segments are shown yellow. The N- and C-termini are represented by green and orange spheres, respectively.
Results
The results indicate that although the rod and twister polymorphs share a bent β-arch architecture, they exhibit distinct inter-protofilament interfaces. While the interface in twister polymorph was formed by the hydrophobic aggregation-triggering non-amyloid-β component (NAC) region from residues G68-A78, the preNAC region from residues E46-A56 comprises the interface in rod polymorph.
The rod polymorph's C-terminal residues were more organized than the twister polymorph, implying higher stability of the rod polymorph than the twister. Nonetheless, six frequent mutations of aS: A53V, A53T, A53E, G51D, H50Q, and E46K, destabilized the rod structure's preNAC interface but did not disrupt the twister polymorph, probably shifting the population from rod to twister.
Visual inspections show that in the presence of the SK9, the aS monomers were more strand-like and extended. Further, the ensemble of the aS in the presence of SK9 shifted toward more solvent-exposed, looser-packed, and larger conformations. This inference implies that the binding of SK9 exposes more hydrophobic residues in aS and probably alters the aS amyloid monomer production by changing the ensemble towards more aggregation-prone conformations.
The interaction of SK9 further augments the selectivity of aS ensembling in the rod-like fibril seeding conformations by inducing higher flexibility, exposure of residues, and a lowered helix-propensity, particularly in the segment E46-A56 that form in the rod fibril polymorph during the inter-protofilament interface. In addition, the interaction between SK9 and the rod fibril substantially enhanced the frequency and lifetime of two contacts, E46-K80 and V52-A76.
Nevertheless, SK9 has little impact on the stability of newly formed or pre-existing rod and twister fibrils. Although there was a stabilization of twister fibril geometry upon the binding of SK9, it does not lead to significant changes in twister fibril quantities.
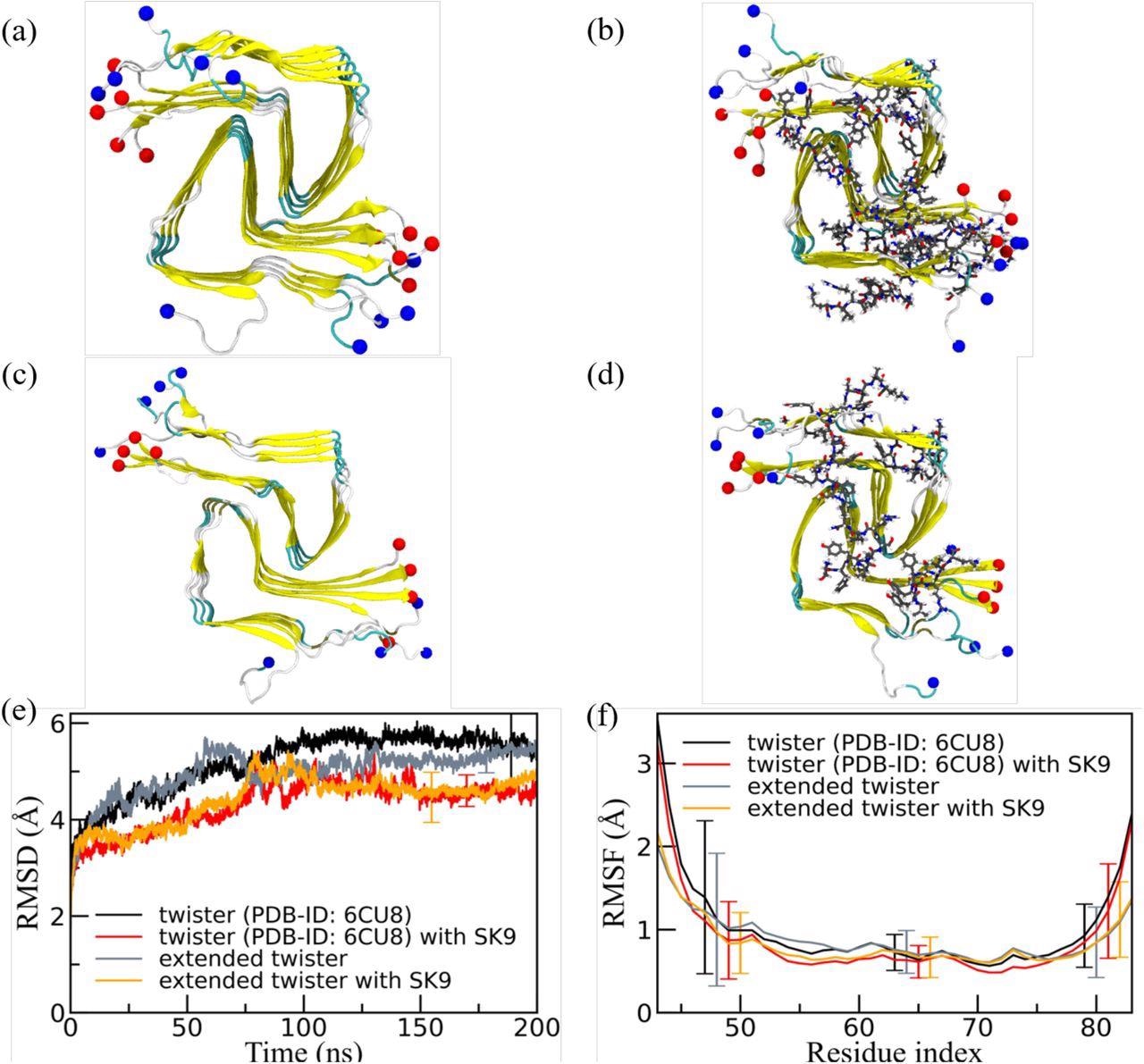
Representative final configurations were extracted from simulations starting from (a) the experimentally determined twister-like α-synuclein fibril model (PDB-ID: 6CU8) and (c) the extended model. Corresponding final snapshots extracted from simulations in the presence of SK9-segment are shown in (b), and (d). N- and C-terminus are represented by blue and red spheres, respectively. Only residues 43-83 are shown for the extended model configurations in (c) and(d). The time evolution of the RMSD in the simulation of these systems is shown in (e), and residue-wise RMSF in (f). We calculate RMSD and RMSF again only for the experimentally resolved region 43-83, i.e., ignoring the disordered and unresolved parts of the fibril models, considering all backbone atoms. Only a few typical error bars are shown to make figures more readable.
Conclusions
The study findings indicate that since the mutations present in aS, which impacted the stability of rod fibrils, were associated with Parkinson's disease, the shift in frequency within rod and twister fibrils will change the likelihood of developing Parkinson's disease. This inference is especially significant during COVID-19 because SARS-CoV-2 SK9 augments the probability of forming rod polymorph.
The study shows that the presence of SK9 alters the ensemble of aS to more aggregation-prone conformations. Interestingly, although the twister fibril was considered more cytotoxic, the interaction of SK9 led to the preference for aS monomer conformations that probably seed the rod-like fibrils, which were associated with a risk for Parkinson's disease.
However, additional simulations employing other amyloidogenic segments of SARS-CoV-2 proteins, specifically in the S, are required to understand whether this effect is selective for SK9. Further, the study indicates the binding of SK9 with the rod and the twister fibril polymorphs had only a minor impact on the stability of these fibrils.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Effect of an amyloidogenic SARS-COV-2 protein fragment on α-synuclein monomers and fibrils, Asis K Jana, Chance W Lander, Andrew D Chesney, Ulrich H.E. Hansmann, bioRxiv 2022.02.21.481360; doi: https://doi.org/10.1101/2022.02.21.481360, https://www.biorxiv.org/content/10.1101/2022.02.21.481360v1
- Peer reviewed and published scientific report.
Jana, Asis K., Chance W. Lander, Andrew D. Chesney, and Ulrich H. E. Hansmann. 2022. “Effect of an Amyloidogenic SARS-COV-2 Protein Fragment on α-Synuclein Monomers and Fibrils.” The Journal of Physical Chemistry B 126 (20): 3648–58. https://doi.org/10.1021/acs.jpcb.2c01254. https://pubs.acs.org/doi/10.1021/acs.jpcb.2c01254.