Mucosal pathogens are responsible for many severe infectious diseases, including influenza, severe acute respiratory syndrome coronaviruses 1 and 2 (SARS-CoV-1 and SARS-CoV-2, respectively), rotaviruses, and gut pathogens like Salmonella or Escherichia coli. The ongoing coronavirus disease 2019 (COVID-19) pandemic has demonstrated the urgency to develop vaccines that can induce both mucosal and systemic adaptive immunity.
Study: Hacking Commensal Bacteria to Consolidate the Adaptive Mucosal Immune Response in the Gut–Lung Axis: Future Possibilities for SARS-CoV-2 Protection. Image Credit: paulista / Shutterstock.com
Introduction
Since the onset of the pandemic, several types of vaccines have been developed. Messenger ribonucleic acid (mRNA) vaccines, for example, have been used to encode the SARS-CoV-2 spike antigen, whereas viral vector vaccines deliver spike RNA into the host cell following vaccination. Others vaccine types include those based on recombinant DNA, inactivated whole virus, and protein subunits.
The next generation of vaccines may be based on reverse vaccinology (RV) and structure-based RV to improve vaccine design using readymade vectors and formulations. Eventually, this may lead to the discovery of better vaccine targets, functional epitopes, and data on viral and vector functions.
Researchers anticipate that these advancements will aid in the development of vaccines targeting more than one epitope with higher vaccine efficacy and the ability to prevent infection with emerging new variants of known pathogens.
Immunoinformatics is another field in which more research is needed to elucidate pathogen antigenicity and immunogenicity using tools like the Immune Epitope Database (IEDB) to identify antigens that bind to major histocompatibility complex (MHC) class I and II. Such technologies within this field can also predict downstream antigen processing, presentation by appropriate cells, and recognition of T- and B-cell receptors, which lead to the activation of antibody-producing cells and effector T-cells.
The Human Microbiome Project is another database that allows interaction networks between host and commensal bacteria to be visualized in greater detail. Commensal bacteria inhabit their own mucosal niches, wherein they build and sustain their microenvironment, as well as regulate host immune responses so that they can coexist. This could help to identify many more targets for RV to allow for commensal microbes to be used to induce innate and cell-mediated adaptive immunity by their interaction with the host mucosa.
In the current review paper, the authors demonstrate how the microbial tools used to ensure homeostasis in the gut microbiome produce mucosal immunity and may lead to the development of bacterial vectors for pathogenic viruses like SARS-CoV-2 for future protective mucosal vaccines.
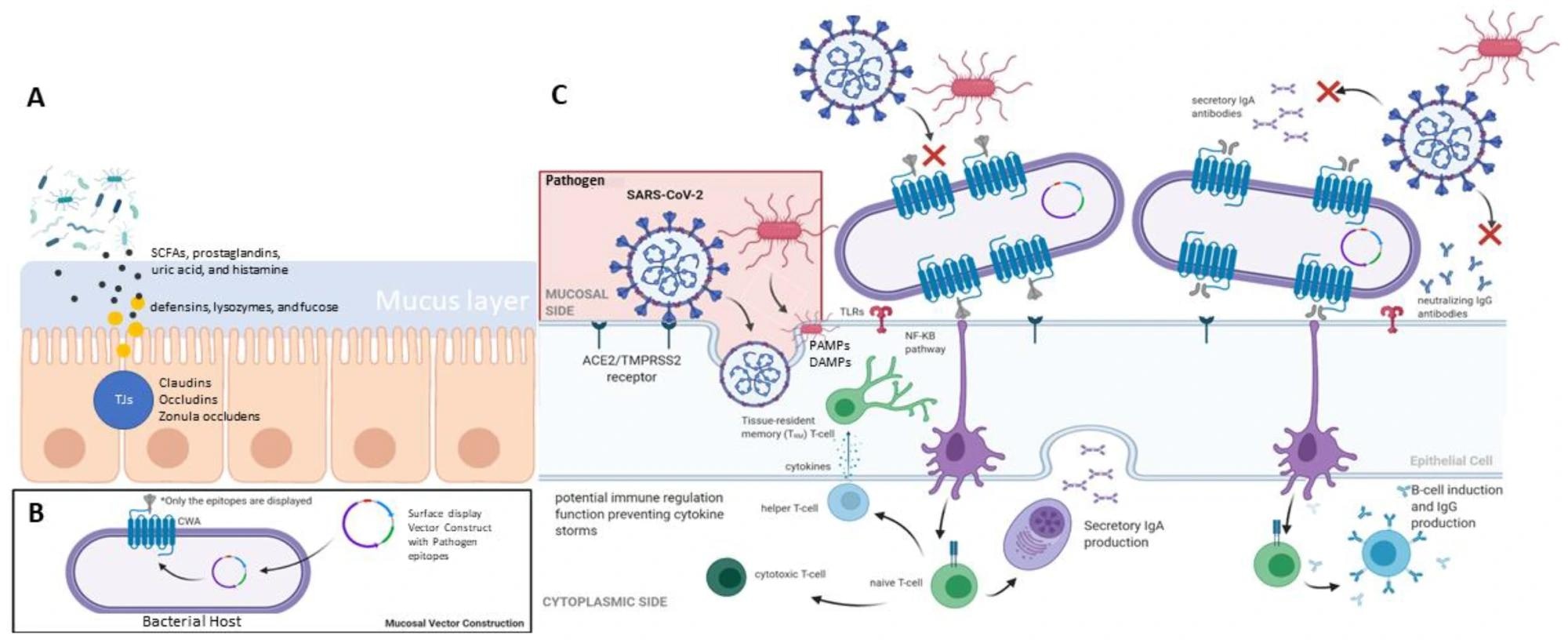 Summary of mucosal-associated immunity in human (A) gut barrier system, (B) mucosal vector construction, and (C) the mucosal response elicited by immune cells.
Summary of mucosal-associated immunity in human (A) gut barrier system, (B) mucosal vector construction, and (C) the mucosal response elicited by immune cells.
Study findings
RV and SBRV have demonstrated the importance of both epitopes and paratopes in the immunogenicity of a given antigen. Targeting immunogenic domains can provide much more effective protection than the whole protein antigen.
Additional components such as liposomes, outer membrane vesicles, and both inorganic and organic adjuvants are often needed to increase the magnitude of the immune response to a given antigen. Attenuated or heat-inactivated bacteria, including bacteria generally regarded as safe (GRAS) such as lactic acid bacteria (LAB), have also been investigated for their adjuvant effects in vaccines.
Since the mucosa-associated lymphoid tissue (MALT) mounts mucosal immune responses, the appropriate MALT must be stimulated to achieve the desired immune response. The different types of MALT tissues include gut-, nasopharynx-, and bronchus-associated lymphoid tissue that is abbreviated as GALT, NALT, and BALT, respectively.
GALT-mediated immunity
The GALT reacts according to the antigen that interacts with intestinal epithelial cells. T regulatory cells (Tregs) with an immunomodulatory effect are generally activated by harmless or beneficial microbes; however, pathogens trigger other signaling pathways through pattern recognition receptors (PRRs). PRRs respond to pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) during antigen-mucosa contact.
At this point, mucosal antigen-presenting cells (APCs) such as dendritic cells (DCs) are activated, along with T- and B-cells and macrophages by these PRRs. The increased activity of CD4+ helper T-cells type 1 (Th1) and Th17 cells also activate APCs and indicate a pro-inflammatory state. These helper T-cells differentiate in response to increasing DC numbers which, in turn, are part of the effort to clear the pathogen as fast as possible.
Immunologic tolerance
When the antigen is harmless, immunologic tolerance can be induced. To this end, immature DCs differentiate into tolerogenic DCs (tolDCs) that stimulate naïve CD4+ T-cells to form Tregs. This leads to inhibition of inflammation in the mucosa, deactivation of effector DCs, and downregulation of Th1 and Th17 cells.
The gamma delta T-cells (γδ T-cells) are another T-cell subtype that responds to common microbial or host antigens that arise during infection, stress, or cancerous changes, in each of which the δ-chain is different. These are CD3+ cells with cytotoxic capabilities, independent of MHC binding.
Secretory IgA
Secretory immunoglobulin A (sIgA) is key to mucosal immunity, as it consists of 80% of total secreted antibodies. These antibodies, which are secreted by polymeric IgA receptors into the body fluid, coat microbes and prevent their contact with the epithelium. While possessing neutralizing and agglutinating activity, sIgA cannot kill bacteria or activate the complement system that destroys bacteria.
New mucosal vaccines based on bacteria may target any of the PAMPs or bacterial metabolites known to induce an immune response, which are also called viability-associated (vita)-PAMPs), provided the PAMP and bacterium belong to the same species.
The NALT and BALT comprise several layers of defense due to the abundance of microbial exposures that occur in these tissues. However, the lungs are sterile and considered to be hyporesponsive to prevent a state of chronic inflammation. This is maintained by the mucociliary clearance system (MCC) and macrophages.
If these fail to cope, the lungs will respond with inflammation.
Earlier bacterial vaccine candidates
Several bacterial vectors have previously been tested, with cell-surface proteins being displayed on carrier proteins that anchor them to the cell. The surface proteins are chosen to stimulate the immune response. Live attenuated bacteria are also reported, including Salmonella typhi, an efficient invader of immune cells, and Listeria monocytogenes.
However, it is crucial to keep in mind the safety issues regarding the use of live-attenuated pathogenic bacteria as a recombinant delivery system due to their potential for reversion to the pathogenic state, which requires further studies.”
Instead, commensal bacteria have been studied, as they offer both safety and immunogenicity. These include Bacillus subtilis, which can induce higher levels of antibodies and neutralizing activity than other species.
Lactobacillus species
Lactococcus lactis (L. lactis) is a starter often used to ferment yogurt and some types of cheese. Recombinant strains of L. lactis that can resist bile acids and gastric acid have been tested as vaccine vectors.
Lactobacilli are even more tolerant to gut conditions, as they can survive for more than seven days. Thus, these species are being studied as possible adjuvants for vaccines against rotavirus, cholera, and Salmonella, as well as pneumonia, anthrax, SARS, and influenza.
Several strains, including L lactis, L. casei, L. plantarum, and L. helveticus have been experimentally demonstrated to produce a robust immune response to many infections.
Importantly, opposition to the use of genetically modified organisms (GMOs) is present, as the engineered genetic material may introduce unwanted phenotypes and inter-bacterial gene transfer could alter the ecosystem markedly. However, with the use of mRNA and viral vector vaccines in the ongoing pandemic, the potential to use lactobacilli to carry SARS-CoV-2 and other mRNA sequences are being seized, despite known challenges with engineering Gram-positive bacteria.
Another promising commensal candidate vector species is Bifidobacterium, which is found in the healthy infant gut, and with probiotic properties. However, this species is strictly anaerobic, thus hindering its use.
Streptococci
Streptococci are commonly found organisms that have a potential role as mucosal vectors for vaccines. Some recent papers have used Streptococcus salivarius to recolonize the upper respiratory tract, thereby reducing the over-abundance of pathogens that cause acute otitis media and decreasing its incidence. Other Streptococcus species like Streptococcus mitis (S. mitis) have been shown to provide protection against pneumococcal infection.
The gut-lung axis is being widely studied, as gut microbiome changes have been recognized to accompany severe COVID-19. Thus, scientists have used commensal bacteria to design COVID-19 vaccines.
Conclusions
While all the studies have been conducted in mice and other rodents, their findings emphasize the need to continue research in this field. Thus, future research may include bacteria colonizing the airways, which are better adapted to this region of the body and would ensure a more robust and long-lived immune response.
It will be necessary to understand the microbiome in detail, including pathogenic species in the community. Protective commensals should be identified, including those which produce antiviral proteins and those which modulate host immunity, as these can maximize vaccine responses.
Such efforts may be helpful in designing COVID-19 vaccines, especially since SARS-CoV-2 will likely remain in circulation for several years. Thus, increasing vigilance and observation of current vaccines, as well as looking for newer methods to strengthen natural mucosal and systemic immunity, is necessary.
Novel approaches are now underway to design universal Sarbecovirus vaccines using structure-guided vaccine designs for SARS-CoV-2 variants, which will achieve broader B or T cell responses to end the current pandemic and prevent emerging new variants.”
Journal reference:
- Pereira, M., Oh, J. K., Kang, D., et al. (2022). Hacking Commensal Bacteria to Consolidate the Adaptive Mucosal Immune Response in the Gut–Lung Axis: Future Possibilities for SARS-CoV-2 Protection. BioTech. doi:10.3390/biotech11010003.