Since their discovery in 2006 by Takahashi and Yamanaka, induced pluripotent stem cells (iPSCs) have seen a remarkable surge in their application within academic and industrial research.
Generated by reprogramming terminally differentiated somatic cells using four essential transcription factors (Oct4, Sox2, KLF4, and c-myc), human iPSCs can proliferate endlessly and differentiate into all major cell types in the human body.
These cells offer immense potential for numerous therapeutic research areas, including cancer, regenerative medicine, and personalized treatment. Moreover, they address various ethical concerns and sourcing challenges associated with pluripotent cells, such as human embryonic stem cells.
Despite notable technological advancements and improved accessibility for researchers, the cultivation of iPSCs continues to present challenges, restricting their widespread application in therapeutics and research. Some common issues include:
- iPSC lines are sensitive and easily perturbed, requiring constant maintenance.
- Poor culture conditions and cell line instability can lead to spontaneous differentiation and loss of pluripotency.
- Generating monoclonal cell lines is challenging, with low efficiency and a lack of proven clonality.
- The labor, cost, and reagent burden associated with iPSC maintenance and workflows is incredibly high and often prohibitive for the end user.

Image Credit: Cell Microsystems
Key highlights
- Viability during single-cell iPSC workflows is improved using the CellRaft Array that provides flask-like culture conditions while maintaining spatial segregation.
- iPSCs can be imaged at the single-cell stage and monitored for clonal growth and pluripotency using software-guided analytical tools.
- CellRaft Technology provides a fully automated, single-instrument iPSC workflow that decreases cost and time per clone while screening 500X more cells per consumable than a standard 96-well plate.
Hypothesis
The inefficient cloning of iPSCs poses a common bottleneck in stem cell research. The hypothesis suggests that the CellRaft Array's favorable culture environment would greatly enhance single-cell iPSC cloning. This enhancement would increase the number of generated clones and reduce the time required for developing a validated cell line.
Materials and methods
To adapt a stable line of iPSCs from aggregate to single-cell passaging, the cells were seeded on the CellRaft Array in a 5:1 ratio of cells to rafts, using 3 mL of iPSC culture media.
To compare limiting dilution and CellRaft Technology, three plates were seeded with the same cell population at a concentration of 10 cells/mL in 100 uL of iPSC culture media, using the Countess II system (Thermo Fisher). The media in the Array was supplemented with a ROCK inhibitor, and half-changes were performed until isolation.
On the day of isolation, the plates were precoated with an extracellular matrix and pre-loaded with media. The CellRaft Array was filled with 5 mL of media. Tra-1-60 (Thermo Fisher), used at the recommended concentration by the manufacturer, was applied to stain the CellRaft Array for one hour.
The CellRaft Array was rinsed and imaged, and positive clonal rafts were selected and isolated into the 96-well collection plate.
Results/Data
Four distinct iPSC cell lines were seeded onto 200 µm CellRaft Arrays coated with either iMatrix-511 (Matrixome), hESC-qualified Matrigel (Corning), or Laminin-521 (STEMCELL Technologies).
As illustrated in Figure 1, individual CellRafts enable easy identification of single iPSCs, providing visual confirmation of their monoclonality. Through consecutive scans on the AIR System, precise monitoring of cell viability, phenotype, and colony formation can be achieved.
The live iPSC colonies were stained with Tra1-60, demonstrating the maintenance of pluripotency on the CellRafts and ensuring the absence of spontaneous differentiation (Figure 2).
Upon forming substantial colonies, CellRafts containing monoclonal iPSC colonies of interest were isolated and transferred into pre-coated 96-well plates with iMatrix-511 or hESC-qualified Matrigel. The outgrowth of colonies of the raft was monitored (Figure 3).
The iPSCs were allowed to continue their growth in the collection plate, resulting in an efficiency of over 70% (Figure 3). Over 200 viable, verified monoclonal iPSC colonies were identified per consumable, in contrast to only 10 colonies per 96-well plate in limiting dilution, which exhibits poor morphology and unknown clonality (Figure 4).
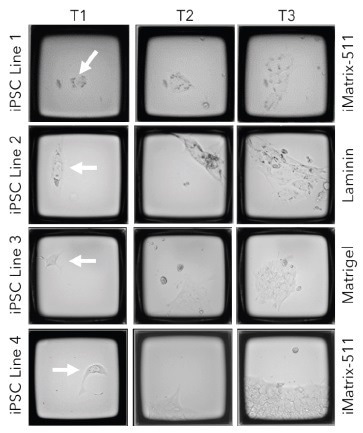
Figure 1. Track and trace of iPSC clone formation on the CellRaft AIR. Four different iPSC cell lines were seeded on CellRaft arrays on one of three coatings (iMatrix-511, h-ESC Matrigel, or Laminin). The arrays were serially scanned starting 4 hours post-cell seeding and every 24 hours after to monitor clone formation. Image Credit: Cell Microsystems
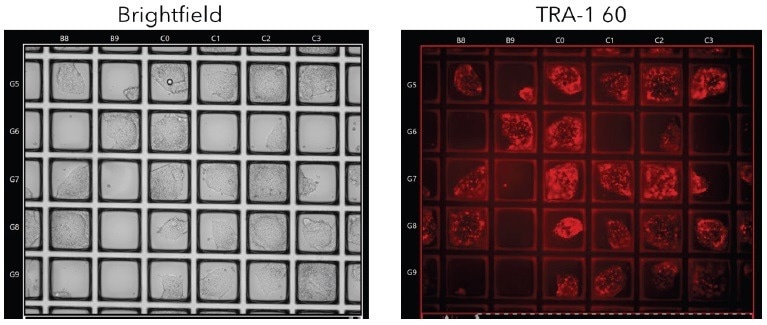
Figure 2. Live Tra-1-60 Staining for Pluripotency. iPSC colonies were stained with live Tra-1-60 antibody to identify pluripotent colonies prior to isolation. Image Credit: Cell Microsystems
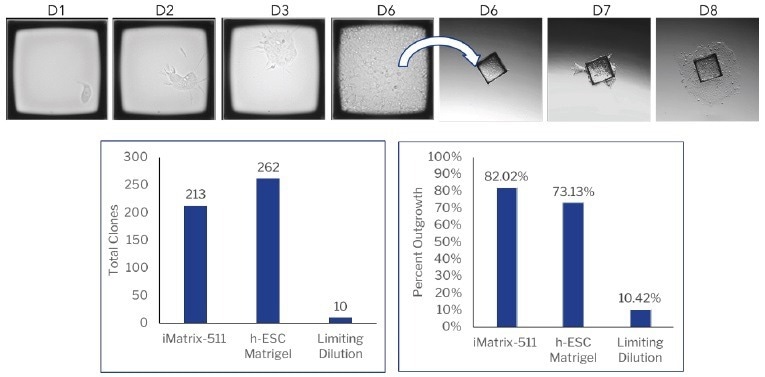
Figure 3. CellRaft AIR vs limiting dilution for monoclonal iPSC development. iPSCs were seeded on either a CellRaft Array coated with iMatrix-511 (Matrixome) or h-ESC Matrigel (Corning) or on iMatrix-511 coated 96 well plates for limiting dilution. Colony formation was monitored using the CellRaft AIR or manual observation. Image Credit: Cell Microsystems
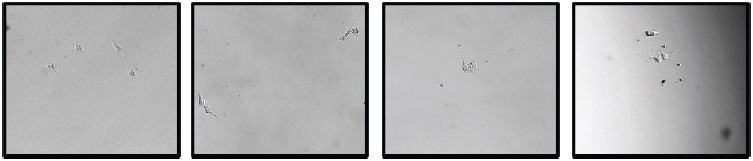
Figure 4. Limiting Dilution images from a 96-well plate with unknown cell clonality. iPSCs were seeded on a pre-coated plate with iMatrix-511 (Matrixome). Image Credit: Cell Microsystems
Discussion
A comparison was made between clonal iPSC development using CellRaft Technology and traditional limiting dilution to evaluate whether these crucial features could overcome bottlenecks that impede iPSC workflows.
Using the CellRaft Array, over 200 single-cell-derived iPSC clones could be generated on a single array, surpassing the 10 clones achieved in a single 96-well plate through limiting dilution.
This workflow necessitated 1000 times less iPSC coating and 2000 times fewer media per cell screened (Table 1).
The acquisition of images over time allows for a comprehensive assessment of monoclonality, ensuring the absence of heterogeneity in successful clones and eliminating the need for downstream clonal characterization, reducing the time required for downstream applications.
Table 1. Consumables per cells screened. Source: Cell Microsystems
| |
iPSCs screened/plate |
uL Coating/cell |
uL Media/cell |
| CellRaft Array |
60,000 |
0.05 |
0.05 |
| 96 well plate |
96 |
52.1 |
104.2 |
Conclusion
The CellRaft Array's capability to offer flask-like culture conditions, while simultaneously preserving the spatial segregation of individual cells, has significantly enhanced the viability of iPSCs and the formation of monoclonal colonies.
A singular consumable can efficiently screen numerous iPSCs, reducing the time needed for cell line generation and therapeutic exploration.
The combination of CellRaft Array technology with the automated imaging and isolation functionalities of the CellRaft AIR System has the potential to augment the usability of iPSCs by streamlining complex workflows involved in iPSC maintenance and cell line development.
About Cell Microsystems
Making innovative tools that enable researchers to image, identify, and isolate viable single cells and clonal colonies
Researchers worldwide in the fields of CRISPR gene editing, oncology, stem cell biology, immunology and neurobiology use Cell Microsystems products, advancing sophisticated discovery across the life sciences.
The company’s CellRaft AIR® System addresses two widespread challenges facing scientists: the ability to actively select viable single cells or clonal colonies based on their phenotype, and match these cells to clonal expansion or molecular analyses. Cells are seeded, imaged, identified, and isolated on Cell Microsystem’s CellRaft® Arrays.
We have tested more than 100 Cell Lines with CellRaft Technology to demonstrate the high outgrowth efficiencies using our platform.
The company currently markets its products to researchers worldwide, and prides itself on being a customer-focused organization responsive to feedback and inspired to fuel deeper contributions to science.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.