By performing a genome-wide CRISPR screen on the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), U.S. researchers identified novel proviral genes and pathways, and highlighted host genes that may regulate coronavirus disease (COVID-19) pathogenesis, as well as revealing potential targets for novel treatment approaches. Their findings are currently available on the bioRxiv* preprint server.
As no vaccines or approved therapeutics are currently available for disease control, the ongoing pandemic of COVID-19 represents a significant risk for public health systems around the world. This can be extended to other emerging coronaviruses, which we have already encountered (such as the original SARS-CoV and MERS-CoV), or the ones that pose a threat for the future.
The entry of SARS-CoV-2 (i.e., the causative agent of COVID-19 disease) into the cell represents the first step in its life cycle, which is mediated by the viral spike protein binding to the cell surface receptor angiotensin-converting enzyme 2 (ACE2). The latter is a significant determinant of host range and cell tropism.
However, a second proteolytic event is warranted in order to expose the viral fusion peptide and enable membrane fusion. This can occur at the target cell plasma membrane by transmembrane protease serine-type 2 (TMPRSS2) or in the endosome by Cathepsin L.
Following the fusion of viral membrane, the viral genetic material is released into the cytoplasm of human cells; there, it is translated and establishes viral replication and transcription complexes before assemblage and budding. Still, the host genes mediating these processes remain elusive.
Nonetheless, the identification of host factors that enable the infection process is of utmost importance to inform COVID-19 pathogenesis mechanisms, reveal host susceptibility variations, but also to identify new host-directed therapies with broad efficacy.
Hence, to uncover host genes necessary for SARS-CoV-2 infection and cell death, the researchers from Yale, Massachusetts Institute of Technology, Harvard, Icahn School of Medicine, and the University of Texas generated a completely novel CRISPR library and pursued the first genome-wide CRISPR screen with SARS-CoV-2.
A genome-wide screening endeavor
In order to identify host genes indispensable for cell survival in response to SARS-CoV-2, this group of researchers decided to use the African green monkey cell line Vero-E6, which is highly amenable to SARS-CoV-2 infection and virus-induced cytopathic effects.
Consequently, they have performed two independent genome-wide screening endeavors by using a genome-wide pooled CRISPR library comprised of 83,963 targeting single guide RNAs, with an average of four single guide RNAs per gene, and one thousand non-targeting control single-guide RNAs.
The exact identification of known proviral genes ACE2 and Cathepsin L demonstrated the technical quality of the screen, providing confidence in finding the additional genes with the propensity to regulate SARS-CoV-2 infection.
As a proof-of-principle, the researchers also tested several small molecule inhibitors, which resulted in the identification of three molecules that had the capacity to inhibit SARS-CoV-2 replication and subsequent virus-induced cell death.
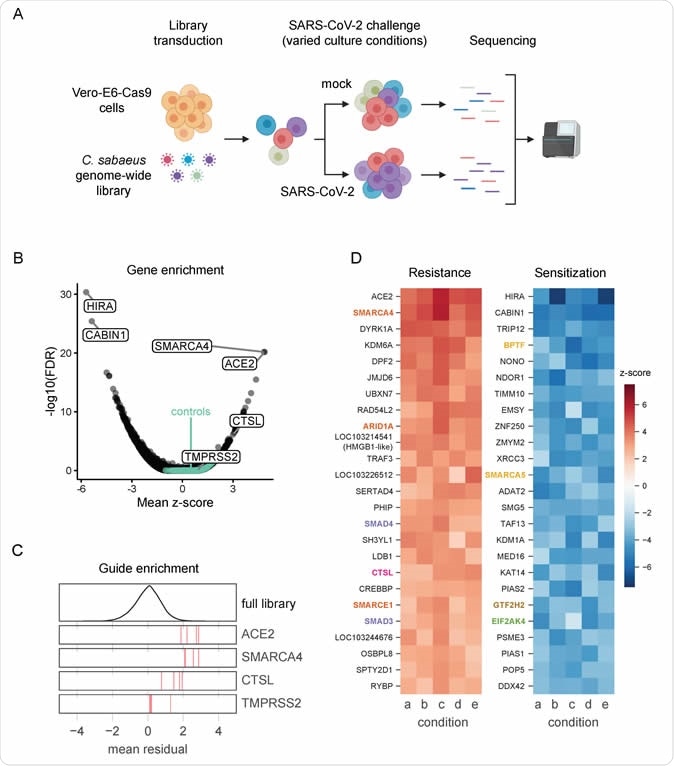
Genome-wide CRISPR screen identifies genes critical for SARS-CoV-2-induced cell death. (A) Schematic of the pooled screen. Vero-E6 cells expressing Cas9 were transduced with the genome-wide C. sabaeus library via lentivirus. The transduced cell population than either received a mock treatment or was challenged with SARS-CoV-2 under various culture conditions. Surviving cells from each condition were isolated and the sgRNA sequences were amplified by PCR and sequenced. (B) Volcano plot showing top genes conferring resistance and sensitivity to SARS-CoV-2. The gene-level z-score and -log10(FDR) were both calculated using the mean of the five Cas9-v2 conditions. Non-targeting control sgRNAs were randomly grouped into sets of 4 to serve as “dummy” genes and are shown in green. (C) Performance of individual guide RNAs targeting ACE2, SMARCA4, CTSL, and TMPRSS2. The mean residual across the five Cas9-v2 conditions is plotted for the full library (top) and for the 4 guide RNAs targeting each gene. (D) Heatmaps of the top 25 genes hit for resistance and sensitivity, ranked by mean z-score in the Cas9-v2 conditions. Genes that are included in one of the gene sets labeled in (Fig 2A) are colored accordingly. Condition a: Cas9v2 D5 2.5e6 Hi-MOI; b: Cas9v2 D5 5e6 Hi-MOI; c: Cas9v2 D2 5e6 Hi-MOI; d: Cas9v2 D10 5e6 Hi-MOI; e: Cas9v2 D5 2.5e6 Lo-MOI.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A panoply of genes involved in SARS-CoV-2 infection
"We discovered genes involved in diverse biological processes including chromatin remodeling, histone modification, cellular signaling, and RNA regulation," study authors emphasize their main findings in the paper available on bioRxiv.
The researchers have assessed 25 of these genes with individual single-guide RNAs in an arrayed format, including both proviral and antiviral genes, and pinpointed small molecule antagonists that provide protection against infection with SARS-CoV-2 and subsequent cell death.
In short, the researchers identified already known SARS-CoV-2 host factors – including the receptor ACE2 and protease Cathepsin L – but also novel proviral genes and pathways. The recent discoveries consisted of one specific chromatin remodeling complex and crucial components of the transforming growth factor beta signaling pathway, which belong to the superfamily of multifunctional cytokines.
"Interestingly, we did not detect several genes previously implicated in SARS-CoV-2 infection including TMPRSS2, PIKFyve, and TPC2 suggesting that these genes are either not essential in a Vero cell-intrinsic manner or that functional redundancy exist with other genes", further explain study authors.
Study authors also unveiled alarmin high-mobility group box 1 (HMGB1) as a key component for SARS-CoV-2 replication; conversely, the loss of histone H3.3 chaperone complex primed the infected cells to virus-induced cell death.
Unveiling potential treatment targets
By highlighting an array of proviral and antiviral host genes, this study has substantial implications for our understanding of COVID-19 pathogenesis, treatment, and vaccine design – most notably by revealing potential therapeutic targets for SARS-CoV-2.
More specifically, genes and pathways that were identified in this study may explain the variation of disease presentation, which may positively correlate with resistance genes, but negatively correlate with sensitization genes on cellular, tissue, and organism level.
"It is intriguing to speculate that the chromatin and histone-modifying genes identified here contribute to the expression of a heterogeneous proviral gene expression program that potentially regulates ACE2 and other viral interacting genes", say study authors.
In conclusion, this study represents the first genome-wide genetic screen performed with any coronavirus, with broadly applicable findings that may prompt the development of host-directed treatments against current and future pandemic strains of coronaviruses.
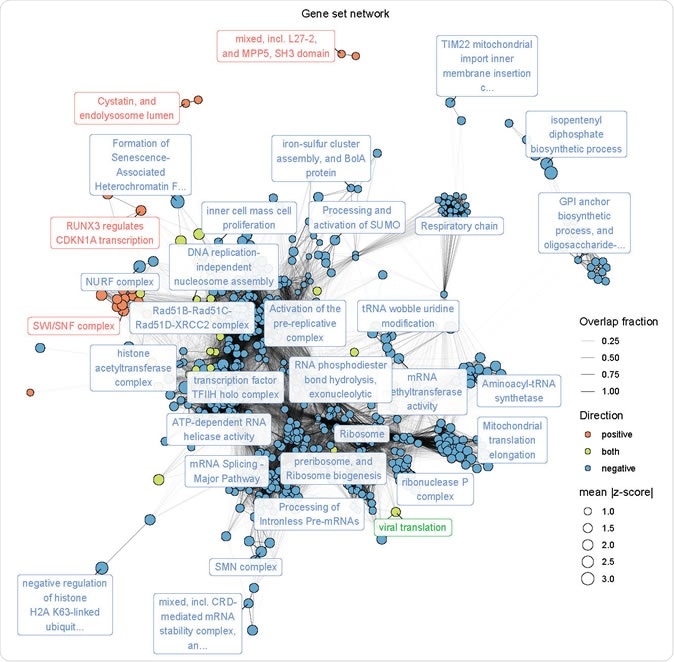
The network of gene sets. Nodes represent significantly enriched gene sets. The size of each gene set is proportional to its mean absolute z-score. Gene sets are colored by the direction in which they score. Edges represent a significant overlap between gene sets. The transparency of each edge is proportional to the fraction of genes shared by two gene sets. Gene sets were clustered using the infomap algorithm and the most central set by PageRank is labeled for each cluster. The Fruchterman–Reingold algorithm was used to lay out the network.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources