The technique, called FD-seq, is a high-throughput method for droplet-based sequencing of single cells compatible with paraformaldehyde (PFA) treatment.
Currently, single-cell sequencing studies that require protein staining or pathogen inactivation are limited by the fact that high-throughput methods are not compatible with PFA treatment, a widely used technique for tissue/cell fixation.
The new FD-seq technique presented by Savaş Tay (The University of Chicago) and colleagues is particularly useful for studying rare subpopulations of cells that require intracellular protein staining and Fluorescence-Activated Cell Sorting (FACS)-enrichment.
In the current study, the team uses FD-seq to address two important problems in virology.
Firstly, the researchers investigated the host factors that support the reactivation of Kaposi’s sarcoma-associated herpesvirus (KSHV) in tumor cells.
Secondly, they used FD-seq to explore the host response in an in vitro model of coronavirus OC43 infection.
This coronavirus is a relative of severe acute respiratory coronavirus 2 (SARS-CoV-2), the agent that causes COVID-19, thereby pointing to the potential use of FD-seq for research into the current pandemic.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
Challenges faced in Single-cell RNA sequencing studies
Single-cell RNA sequencing (scRNA-seq) has important wide-ranging applications, from discovering new cell types to mapping the transcriptome of embryonic stem cells.
Droplet-based scRNA-seq technologies are incredibly powerful, owing to their high throughput, which enables the analysis of thousands of single cells in a single experiment.
However, even with the availability of these high-throughput techniques, the study of rare cell populations remains challenging because it often requires protein-based enrichment of the cell population.
This enrichment can sometimes be achieved using cell surface markers and FACS. Still, some cell types require intracellular protein staining, and this often means fixation with PFA is needed to increase the signal-to-background ratio.
However, PFA fixation presents a challenge in RNA sequencing because the nucleic acids are chemically cross-linked to intracellular proteins.
“In order to retrieve high-quality RNA from single PFA-fixed cells, an appropriate cross-link reversal protocol that maintains RNA integrity and minimizes RNA loss is crucial,” said Tay and colleagues.
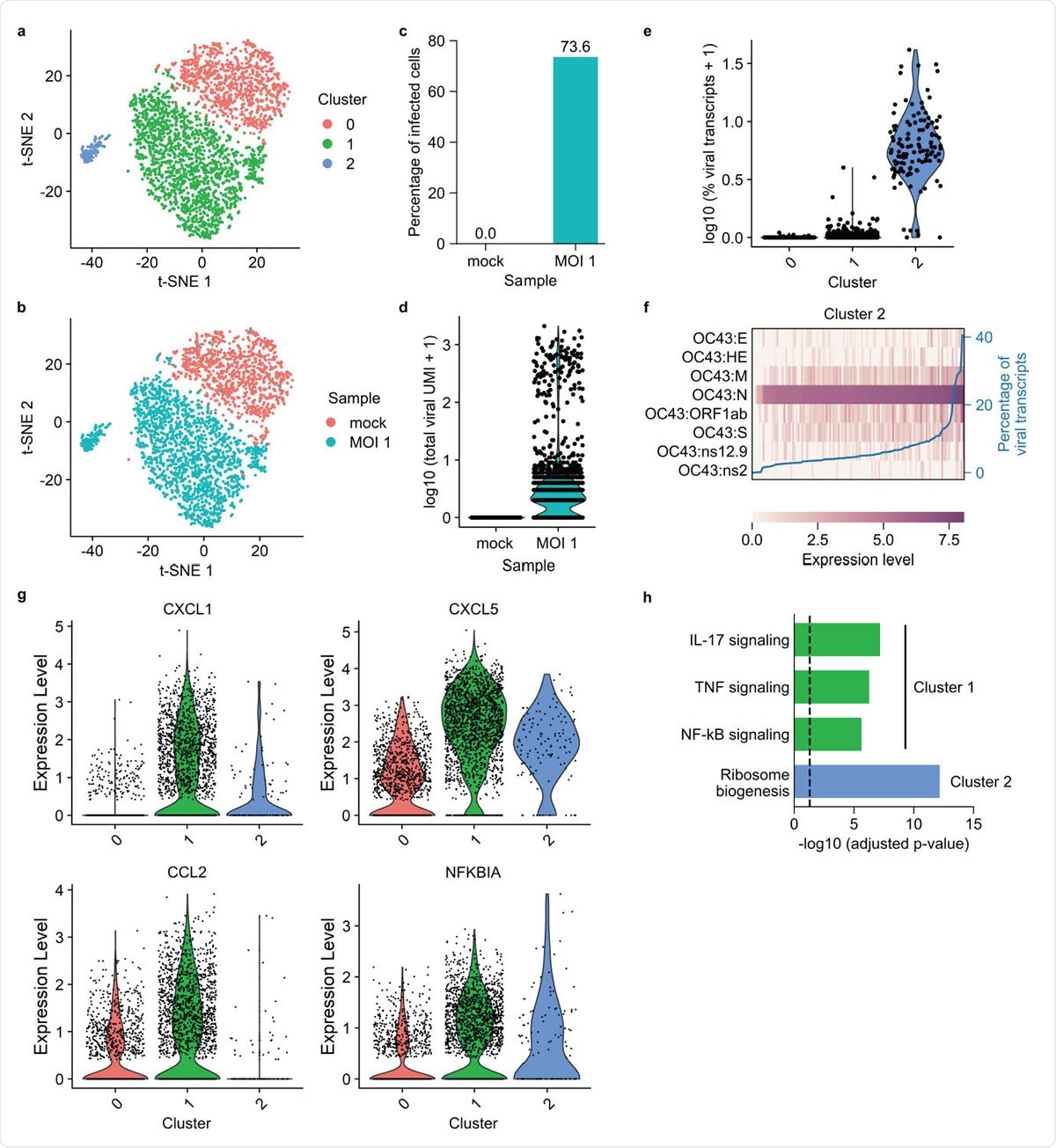
FD-seq reveals pro-inflammatory bystander cells after coronavirus OC43 infection. (a-b) t-SNE plots with cells colored by (a) cluster identity or (b) sample type. (c) Bar plot showing the percentage of infected cells (defined as cells that expressed at least 1 viral transcript) of mock infected and MOI 1 sample. (d) Violin plot showing distribution of total viral transcript counts. (e) Violin plot showing the distribution of the percentage of total viral transcript by cluster identity. (f) Heatmap showing the relative expression level of each viral gene of each single cell in cluster 2. The blue line shows each cell’s percentage of total viral transcripts. (g) Violin plots showing the expression of four representative immune-related genes that are upregulated in cluster 0. (h) Bar plot showing the adjusted P-values of upregulated KEGG pathways in clusters 1 and 2. The vertical dashed line indicates P-value = 0.05.
What did the researchers do?
Now, the team has shown that FD-seq is a particularly useful high-throughput method for droplet-based RNA sequencing of rare cell subpopulations that require PFA fixation.
The team used the method to address two challenging problems in virology.
First, they investigated the host factors that influence Kaposi’s sarcoma-associated herpesvirus (KSHV) reactivation in tumor cells.
The researchers say there is considerable interest in understanding the molecular details of the host factors that modulate KSHV latency and reactivation because these are known to contribute to viral tumorigenesis. Therapeutic induction of reactivation could sensitize latently-infected cells to currently available anti-herpesvirus drugs.
By using FD-seq to analyze a rare population of cells that support KSHV reactivation, the team identified four host genes - CDH1, CORO1C, ISCU, and TMEM119 - whose levels strongly correlated with viral reactivation.
Using live-cell imaging and a time-course study of viral gene expression, the researchers found that the TMEM119 (transmembrane protein 119) gene had the most pronounced effect in terms of inducing KSHV reactivation. Further analysis by bulk RNA-seq indicated that this effect of TMEM119 might be mediated through the MAPK signaling pathway.
Applying the technique to a relative of SARS-CoV-2
Next, the researchers investigated the immune response of human lung cells to infection with coronavirus OC43, a relative of SARS-CoV-2 that causes the common cold.
“OC43 has been successfully used to discover drugs that inhibit SARS-CoV-2 replication in vitro,” says the team.
Following exposure to the virus, most cells were unable to support a high level of viral gene expression and instead upregulated pro-inflammatory immune signaling pathways involving cytokines such as interleukin- 17 and tumor necrosis factor.
This finding points to the possibility that the excessive cytokine release or “cytokine storm” that has been associated with severe COVID-19 could be driven by bystander cells and that these cells could serve as a potential therapeutic target.
“We showed a potential application of FD-seq for investigating the host response in a coronavirus infection in vitro model, which also suggests its possible use in COVID-19 pandemic research,” writes the team.
A useful technique for future studies
The researchers conclude that FD-seq is a valuable tool for integrating protein activity with transcriptome information.
“Transcription factor expression levels and phosphorylation can be integrated with whole transcriptome analysis by combining intracellular protein staining with FD-seq,” they write.
Furthermore, “in future studies, FD-seq could potentially be improved for sequencing formalin-fixed paraffin-embedded (FFPE) tissues, the achievement of which would enable high-throughput single-cell sequencing of readily available samples in tissue banks,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources