Researchers have recovered and identified the various types of neutralizing antibodies for SARS-CoV-2 from convalescent COVID-19 patients. Of these, they identified three that are extremely potent and could be used to develop treatment therapies.
As the COVID-19 pandemic sweeps across the globe, there have been urgent efforts to contain its spread. In the absence of any drugs or vaccines against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the strategy has been to implement social distancing and quarantine.
However, these strategies have not been efficiently implemented in all countries, limiting disease prevention. Thus, the need to develop vaccines and therapies to combat the disease has become critical.
It has also been predicted that the disease will continue to spread and there will be further waves until we reach herd immunity, either using a vaccine or naturally. Thus, there is a need to develop treatments for the disease in addition to developing vaccines.
Several types of human monoclonal antibodies (mAbs), which are antibodies produced in the lab to target a specific antigen, are now commercially available for treating a variety of diseases. Given the significant clinical experience in making mAbs, mAbs to target SARS-CoV-2 may be a route to develop therapies quickly.
Several researchers have been isolating human mAbs from COVID-19 patients who have recovered. Many of these have only moderate potency to neutralize the virus. A few grams of the antibodies may be required per person to effectively combat the virus, which will require injecting the antibodies in the bloodstream, rather than intramuscular injections.
Human monoclonal antibodies from recovered patients
A new study published on the preprint server bioRxiv* reports on screening and characterizing mAbs from recovered COVID-19 patients to identify and recover the most potent antibodies against SARS-CoV-2.
The SARS-CoV-2 virus infects humans by attaching itself to the human angiotensin-converting enzyme 2 (ACE2) via the spike protein. The spike protein exists in a metastable conformation before fusion and a stable conformation after fusion to ACE2. The receptor-binding domain (RBD) of an S1 subunit of the spike protein attaches to ACE2.
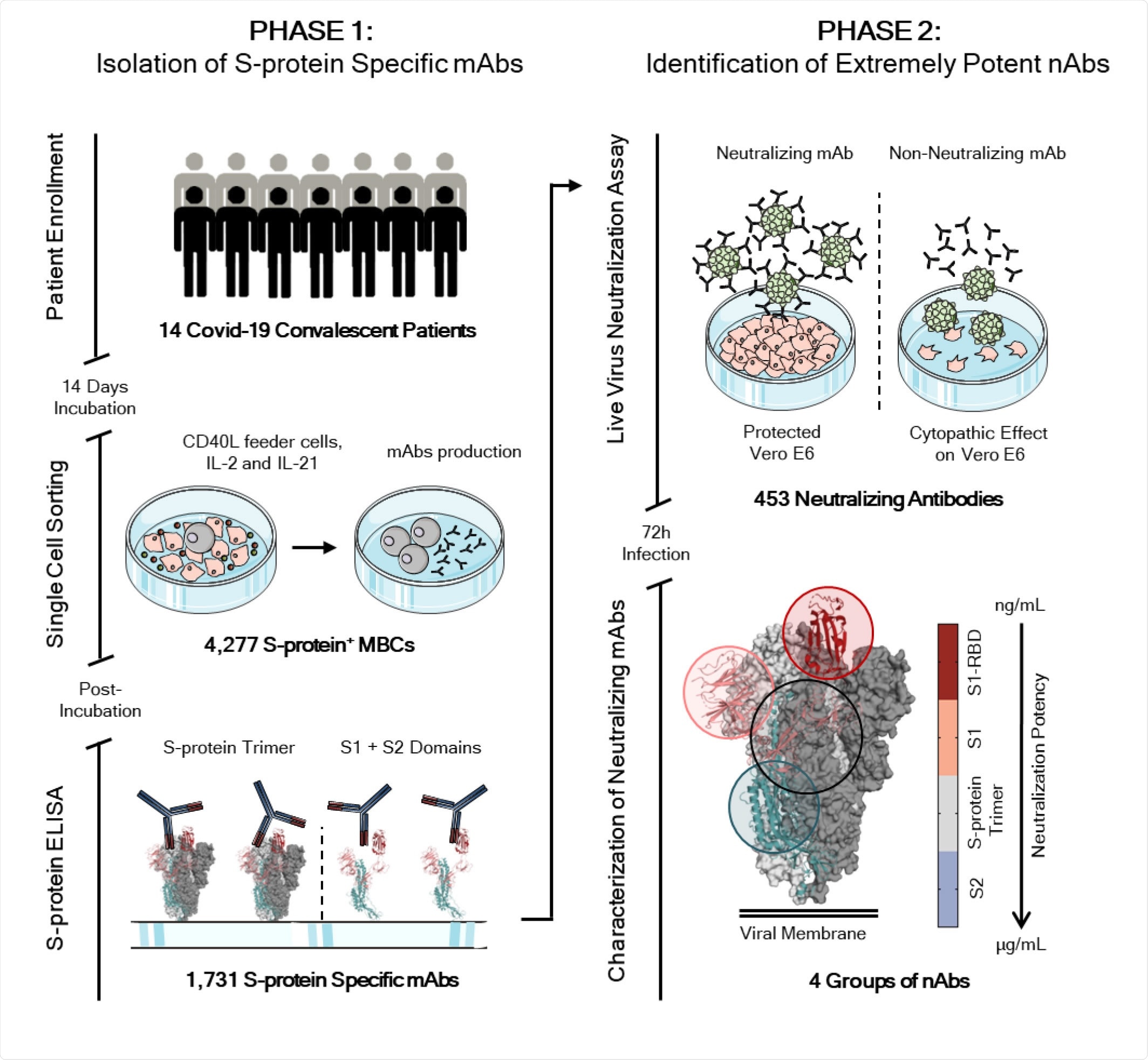
Workflow and timeline for SARS-CoV-2 neutralizing antibodies identification. The overall scheme shows two different phases for neutralizing antibodies (nAbs) identification. The phase 1 consisted in the enrolment of Covid-19 patients (14) from which PBMC was isolated. Memory B cells were single cell sorted (N= 4,277) and after 2 weeks of incubation the antibodies screened for their binding specificity against the S-protein trimer and S1/S2 domains. Once S-protein specific monoclonal antibodies were identified (N=1,731) the phase 2 started. All specific mAbs were tested in vitro to evaluate their neutralization activity against the live virus and 453 nAbs were identified. nAbs showing different binding profiles on the S-protein surface were selected for further functional characterization.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
So, the researchers targeted mAbs specific to the spike protein. To recover these mAbs, the authors collected peripheral blood mononuclear cells (PBMCs) from 14 convalescent patients. They stained the spike protein to identify the spike protein-specific memory B cells (MBCs). They recovered about 4300 different MBCs, which they incubated for two weeks to allow for the production of immunoglobulins.
After that, they screened these antibodies produced for specific binding to the spike protein using ELISA and recovered 1731 mAbs.
The authors tested the ability of these 1731 mAbs to block the binding of the spike protein to Vero E6 cell receptors and to neutralize SARS-CoV-2 using in vitro assays. They found 19.6% of the mAbs blocked spike protein binding with potency ranging from 50% to 100%.
When they tested the ability of these mAbs to protect the Vero E6 from SARS-CoV-2 infection, they found 453 mAbs neutralized the live virus.
Most of the neutralizing mAbs, about 58%, specifically recognized the S1 subunit, about 7% specifically recognized the S2 subunit of the spike protein, and about 35% recognized the spike protein trimeric configuration.
Next, the authors tested how good was the neutralizing potency of the mAbs identified above. They found about 66% had low neutralizing potency, requiring more than 500 ng/mL for achieving IC100, about 24% had medium potency, and about 1.4% were extremely potent, with an IC100 of less than 10 ng/mL.
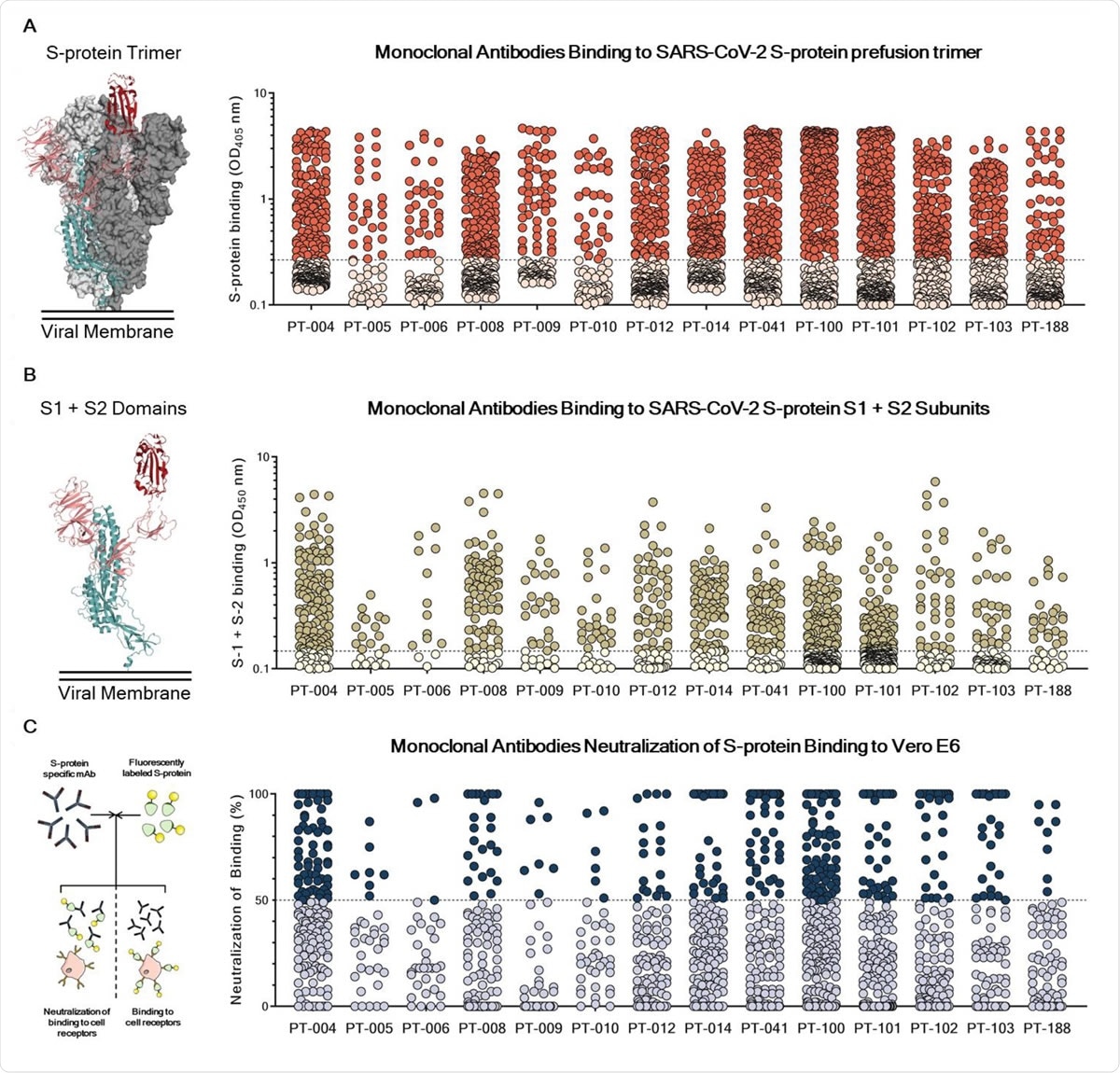
Identification and characterization of SARS-CoV-2 S-protein specific mAbs. (A) The graph shows supernatants tested for binding to the SARS-CoV-2 S-protein stabilized in its prefusion conformation. Threshold of positivity has been set as two times the value of the blank (dotted line). Red dots represent mAbs which bind to the S-protein while pink dots represent mAbs which do not bind. (B) The graph shows supernatants tested for binding to the SARS-CoV-2 S-protein S1 + S2 subunits. Threshold of positivity has been set as two times the value of the blank (dotted line) Darker dots represent mAbs which bind to the S1 + S2 while light yellow dots represent mAbs which do not bind. (C) The graph shows supernatants tested by NoB assay. Threshold of positivity has been set as 50% of binding neutralization (dotted line). Dark blue dots represent mAbs able to neutralize the binding between SARS-CoV-2 and receptors on Vero E6 cells, while light blue dots represent non-neutralizing mAbs. The total number (N) of S-protein specific supernatants screened by NoB assay is shown for each donor.
Extremely potent antibodies
The researchers selected 14 mAbs from their first round of screening to continue investigating. Of these, some were able to bind to the S1 or S2 subunits and others to the spike protein trimeric configuration.
Of the neutralizing mAbs binding to the S1 subunit, 6 mAbs bound to the RBD and were extremely potent for neutralizing the virus.
Antibodies that bound to the spike protein trimeric configuration had lower neutralizing potency compared to those that bound to RBD or the S1 subunit. The antibodies binding only to the S2 subunit showed the lowest neutralizing potency.
“From these data we conclude that in convalescent patients most of the observed neutralization titers are mediated by the antibodies with medium-high neutralizing potency,” write the authors. The extremely potent antibodies and antibodies binding to S2 are too few to have any effect.
Hence, it will be important to understand the types of antibodies produced after vaccination, write the authors, as vaccination may produce different sets of neutralizing antibodies.
Of the 453 mAbs the authors screened, the team found three mAbs that had extremely high neutralization potency against the SARS-CoV-2 strain from Wuhan and the D614 strain.
These antibodies will allow the use of small quantities to be used to achieve therapeutic benefits, reducing the cost of treatment.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources