The coronavirus disease (COVID-19) pandemic is far from over. It has spread across the globe, infecting more than 46.5 million people.
In October 2020, an outbreak of at least 50 COVID-19 cases was reported surrounding individuals employed at or visiting the White House.
There have been over 9.2 million confirmed cases in the United States alone, and over 230,000 deaths are reported. COVID-19 has been repeatedly associated with localized outbreaks surrounding social settings like weddings and bars as well as workplaces, including so-called "superspreader" events.
.jpg)
President Donald J. Trump announces Judge Amy Coney Barrett as his nominee for Associate Justice of the Supreme Court of the United States Saturday, Sept. 26, 2020, in the Rose Garden of the White House. (Adapted from Official White House Photo by Amy Rossetti)
Numerous high-profile individuals were infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), including President Donald Trump, who was hospitalized for three days. The event has been tagged as the most significant health crisis for a president in nearly four decades. Trump has since recovered; however, some questions remain, where did the virus come from, when did it arrive at the White House, and how was it spread?
Now, a new study revealed the genetic signature of the SARS-CoV-2 that spread in the White House and most likely infected President Trump. The team applied genomic epidemiology to shed light on the origins of the outbreak.
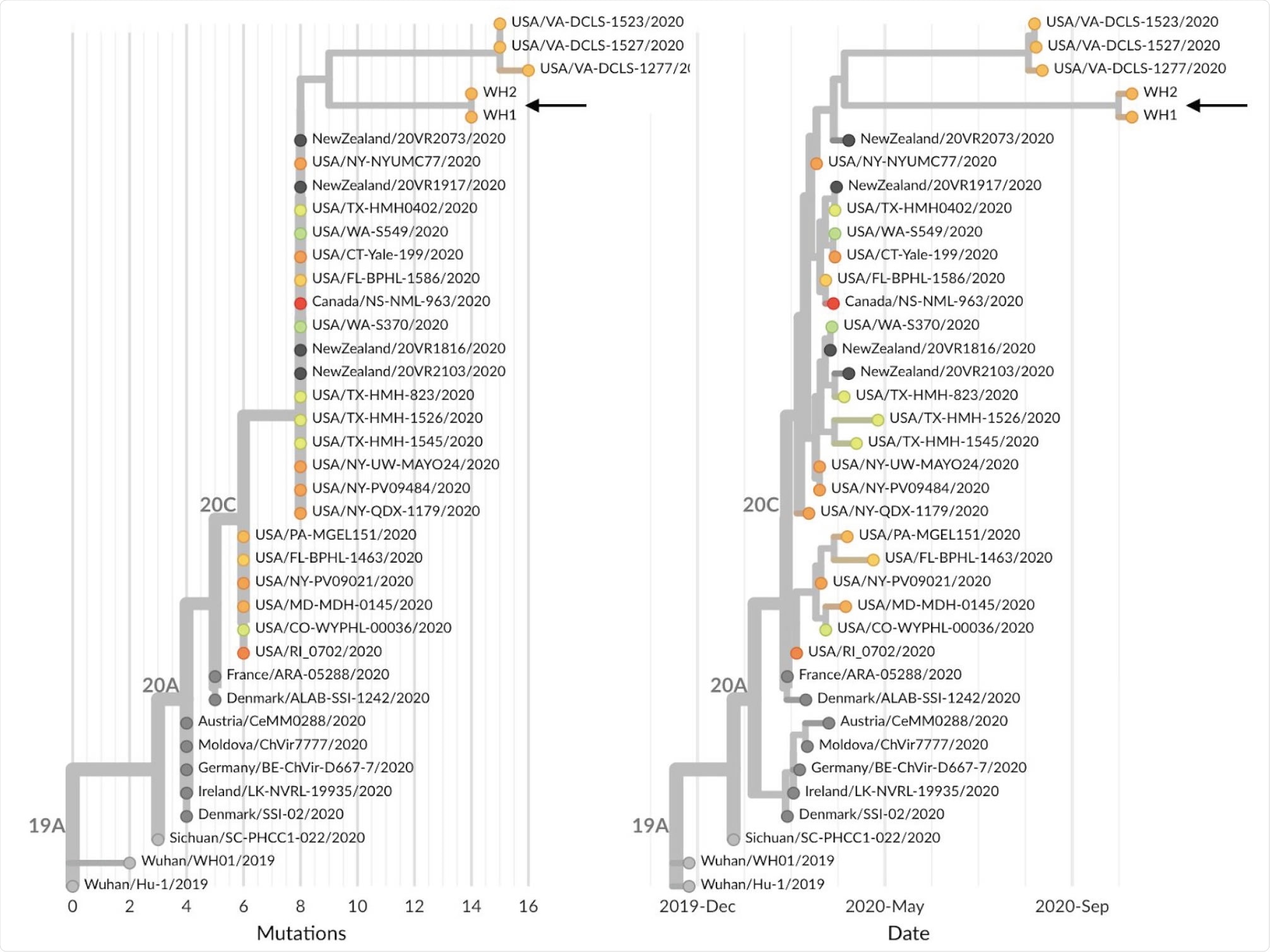
Phylogeny of 38 SARS-CoV-2 viruses that are either sister lineages to WH1 and WH2 or directly ancestral in the global maximum-likelihood phylogeny. Shown are both (A) phylogeny with branch lengths scaled by number of mutations from Wuhan reference genome and (B) temporally resolved phylogeny with branch lengths estimated according to a molecular clock analysis. Both panels are colored according to state of sampling for US samples or colored gray if samples were from outside the US. An interactive version of this figure is available at nextstrain.org/community/blab/ncov-wh/lineage
The White House outbreak
At 12:54 a.m. EDT on October 2, 2020, President Trump tested positive for SARS-CoV-2 infection and was hospitalized at Walter Reed National Military Medical Center in Bethesda, Maryland. However, just days after, he was discharged and allowed to return to the White House. Where Trump contracted the virus remains a mystery. Still, much of the focus has been on a September 26 event in the Rose Garden, where the president announced his nomination of Judge Amy Coney Barrett for the vacancy on the Supreme Court.
The event, which had at least a dozen guests, has been dubbed a super spreader event. Further, debates commenced on whether the White House has done enough to trace guests and their contacts.
By October 7, the Federal Emergency Management Agency memo revealed that 34 White House staff members, housekeepers, and other contacts had contracted the virus. Among the president's contacts, the First Lady, Melania Trump, a Navy admiral, and some campaign aides all tested positive.
At least 50 COVID-19 cases were reported surrounding individuals employed or visiting the White House by October 30.
The study
The researchers at the Fred Hutchinson Cancer Research Center and the University of Washington wanted to determine the viral genome and genetic signature of the virus spread in the White House.
Due to the long incubation periods and a wide range of disease severity, with some people developing no symptoms, contact tracing for COVID-19 spread is difficult. However, the genetic sequencing of the SARS-CoV-2 virus provides an alternative method to investigate its transmission and spread.
The technology aids in determining genetic relationships among sequenced samples. These sequences help make assumptions about how infections are related. Since the coronavirus roughly mutates every two weeks in a transmission chain, it is possible to use patterns of shared mutations to provide a better understanding of the origins of the outbreak in the White House.
To arrive at the study findings, which appeared on the pre-print journal medRxiv*, the researchers applied genetic epidemiology to explore the outbreak's origins.
The team enrolled two people with exposures tied to the White House COVID-19 outbreak and sequenced their infections by collecting nasal swabs. The two study participants, referred to as WH1 and WH2, were not in direct contact with each other before their COVID-19 diagnoses.
The researchers revealed the SARS-CoV-2 genetic relationships between WH1 and WH2. They used a hybrid capture or shotgun metagenomics, resulting in 550X average depth-of-coverage of WH1, getting a consensus genome of 29,857 resolved bases. It also contains 14 mutations relative to the original strain from Wuhan City in China, the Wuhan/Hu-1/2019 (241T, 1059T, 1977G, 3037T, 7936T, 14250T, 14408T, 16260T, 18417C, 19524T, 20402T, 23403G, 25563T, 28821A).
Meanwhile, the WH2 yielded a partial genome with 50X average coverage and 2,643 resolved bases. Since WH2 is a partial genome, the team wanted to characterize the genetic match between WH1 and WH2.
Of the 14 sites that differentiate WH2 from reference, five of these sites had been read coverage in WH2. Hence, the mutations place WH1 and WH2 within circulating genetic diversity in the United States.
By comparing WH1 and WH2 to all sequences across the country, they found that the viruses were descended from viruses samples in the U.S., particularly in Florida, New York, Texas, Connecticut, and Washington, and also from other countries such as Canada and New Zealand. Further, they share characteristics with some mutations, such as the A1977G, G7936T, G14250T, T18417C, C19524T, and C20402T.
"These observations suggest a transmission chain leading to WH1 and WH2 from viruses circulating in the USA in March and April that collects an additional 5 mutations over these 6 months of circulation, consistent with the overall observed rate of molecular evolution of SARS-CoV-2," the team explained.
The team concluded that given their genetic similarity, WH1 and WH2 are closely related. Since these patients said they had no direct contact with each other, they had exposure to events tied to the outbreak in the White House.
"We believe that a shared epidemiological connection through the White House COVID-19 outbreak is the most parsimonious explanation for their infections' genetic similarity," the team added.
The W.H. lineage determined in the study was responsible for the other infections in the White House. The team said that though they know these cases are connected, they are unsure if the virus strain has been circulating more broadly in the D.C. area. Hence, the team suggests broader genetic sequencing, including other cases in the White House.
Source:
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.