By tracing maternal and paternal inheritance without parental DNA, scientists have mapped dozens of parent-specific genetic effects, uncovering how the same variant can drive or protect against disease depending on which parent it comes from.
 Study: Parent-of-origin effects on complex traits in up to 236,781 individuals. Image Credit: isak55 / Shutterstock
Study: Parent-of-origin effects on complex traits in up to 236,781 individuals. Image Credit: isak55 / Shutterstock
In a recent study published in the journal Nature, an international team of researchers developed a novel approach to infer parent-of-origin (PofO) of alleles without parental genomes.
Genome-wide association studies identify additive genetic effects, assuming that the phenotypic effect depends on the number of copies of a given allele, irrespective of their parental origin. Nevertheless, some sequence variants could exhibit distinct phenotypic effects based on whether they are paternally or maternally inherited, a phenomenon known as PofO effects (POEs).
POEs have been traditionally linked to genomic imprinting, wherein only one parental gene is expressed based on its origin. They are believed to occur due to parental conflict over resource allocation to offspring, leading to opposing parental influences. Notwithstanding their significance, POEs are underexplored in complex traits due to the lack of parental genomes.
The study and findings
In the present study, researchers developed a multi-step approach to infer the PofO of alleles in large-scale biobanks without parental genomes. They used pairwise kinship estimates to identify related individuals (parent-offspring duos, trios, and siblings) in the United Kingdom Biobank (UKB) cohort. For more distant relatives (up to fourth degree), they were grouped into two parental sides without differentiating between maternal and paternal sides.
These family groups were referred to as surrogate parents. A validation cohort was created with 2,141 individuals who had actual parental genomes. The identity-by-descent information was used from surrogate parents for inter-chromosomal phasing to identify variant sets co-inherited across and within chromosomes. Mitochondrial DNA (mtDNA) and X chromosome inheritance patterns were examined for persons with second to fourth degree surrogate parents.
For sibling pairs, crossover positions were inferred, and PofO was probabilistically assigned using sex-specific recombination maps. Overall, PofO estimates were obtained for 286,666 individuals. High-confidence estimates (PofO probability ≥0.99) were obtained for over 40% of individuals, with 94% concordance across three predictors (crossovers, mtDNA, and X chromosome).
Further analyses were restricted to 109,385 white British individuals. The overall PofO inference accuracy, estimated using the validation cohort, was 97.94%, with over 80% of individuals exceeding 99% accuracy. Next, the team used the resultant PofO-resolved data to conduct PofO-specific association scans on 59 complex traits and more than 14,000 protein quantitative trait loci (pQTLs).
The team used a multi-purpose approach to uncover novel putative associations. They focused on imprinted regions (within a 500 kb window of known imprinted genes), which are more likely to exhibit POEs. Next, they focused on regions with additive association with the examined trait (additively associated regions). Finally, POEs were scanned genome-wide without prior assumptions. Within imprinted regions, 11 significant POEs and 16 suggestive POEs were detected.
Besides, there was a statistically significant enrichment of POEs in growth and metabolism traits compared to other trait categories (OR=5.35, P=0.018). Over one-third of these POEs demonstrated bipolar effects (opposite parental influences), where alleles inherited from one parent increased trait values but decreased them when inherited from the other parent.
A bipolar effect of rs62471721 was detected on triglyceride levels at the 7q32.2 imprinted region. Two additional variants exhibited suggestive bipolar POEs on sex hormone-binding globulin and high-density lipoprotein cholesterol. Further, three independent POEs were identified at the H19/IGF2 imprinted region affecting standing height. A novel association involved the paternal T allele of rs576603.
This variant is a known expression QTL for the paternally expressed insulin-like growth factor 2 (IGF2) gene and a splice QTL for the maternally expressed H19 gene. Further, the researchers identified significant bipolar POEs of one variant (rs10838787) at the 11p15.5 locus on type 2 diabetes (T2D) – refining a previously reported but unreplicated association – wherein the paternal A allele increased the risk of T2D by ~25% compared to maternal inheritance, while the maternal A allele was protective.
At the same locus, a bipolar POE of rs10838787 on glycated hemoglobin and a suggestive bipolar POE of rs4417225 on glucose with male-specific effects were detected. Further, six significant POEs were detected within additively associated regions. Two POEs were novel associations, with the rs2293607 C allele reducing telomere length more strongly when inherited paternally (exhibiting an 'asymmetric polar' pattern where both parental alleles shorten telomeres but paternal inheritance has more potent effects), while the rs11100479 T allele increases telomere length when inherited paternally.
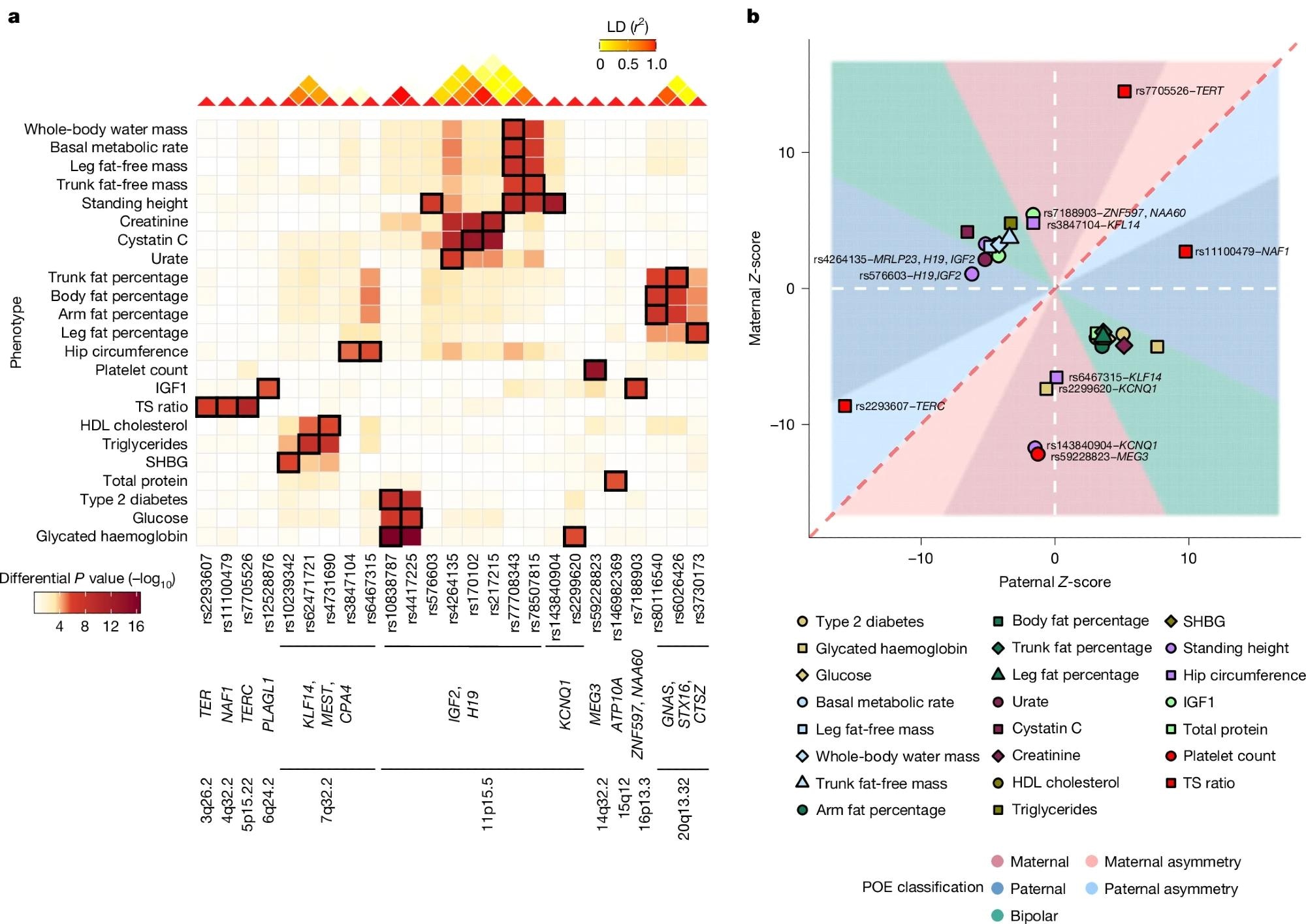 a, Heatmap summarizing the differential P value for all significant POEs identified in this study, computed using REGENIE (two-sided test). Columns correspond to genetic variants ordered by genetic position and annotated with the genes and chromosome bands, whereas rows represent phenotypes. The colour intensity represents the magnitude of the differential P value, with darker shades indicating stronger differential effects. Cells with black rectangles indicate significant POEs identified in this SNP–trait pair. The linkage disequilibrium (LD) heatmap (top) shows the linkage disequilibrium (r2) between the variants. b, Scatter plot illustrating the parental Z-scores for all significant POEs identified in this study. Each point represents a significant POE, coloured and shaped by phenotype. The dashed red line represents the line of equality, and the dashed white lines represent zero values for paternal and maternal Z-scores, respectively. Areas are filled according to the POE classification. Labelled points correspond to POEs classified as paternal or maternal effects. Unlabelled points correspond to bipolar POEs.
a, Heatmap summarizing the differential P value for all significant POEs identified in this study, computed using REGENIE (two-sided test). Columns correspond to genetic variants ordered by genetic position and annotated with the genes and chromosome bands, whereas rows represent phenotypes. The colour intensity represents the magnitude of the differential P value, with darker shades indicating stronger differential effects. Cells with black rectangles indicate significant POEs identified in this SNP–trait pair. The linkage disequilibrium (LD) heatmap (top) shows the linkage disequilibrium (r2) between the variants. b, Scatter plot illustrating the parental Z-scores for all significant POEs identified in this study. Each point represents a significant POE, coloured and shaped by phenotype. The dashed red line represents the line of equality, and the dashed white lines represent zero values for paternal and maternal Z-scores, respectively. Areas are filled according to the POE classification. Labelled points correspond to POEs classified as paternal or maternal effects. Unlabelled points correspond to bipolar POEs.
Further, the team tested whether loci with POEs on obesity-related traits and adult height exhibit POEs on these traits in early life. To this end, they used the childhood data in the UKB and longitudinal body mass index (BMI) and height data collected at 11 time points up to age 8 in the Norwegian Mother, Father, and Child Cohort study (MoBa). Two adult height loci (rs576603 and rs77708343) had significant POEs on comparative height size at age 10 in the UKB.
The bipolar POE of rs77708343 also showed associations with infant height across all time points in MoBa. These effects mirrored those observed in adulthood, suggesting POEs influence early growth trajectories from infancy. Further, rs6467315 exhibited POEs on infant BMI at seven time points in MoBa. The maternal G allele of rs6467315 was associated with a higher BMI in infancy and lower hip circumference and BMI in adulthood – a reversal effect observed across development. Critically, these effects were not attributable to maternal rearing influences when analyzing untransmitted alleles.
Four significant POE pQTLs were identified from over 14,000 known pQTLs. POEs at carboxypeptidase A4 (CPA4) – where the lead POE variant differed from the top additive pQTL – and delta-like non-canonical notch ligand 1 (DLK1) aligned with their known imprinting, whereas POEs at period circadian regulator 3 (PER3) and ADAM metallopeptidase domain 23 (ADAM23) exhibited paternal-specific effects, suggesting potential incomplete or context-dependent imprinting beyond established regions.
Finally, to replicate POEs, a similar multi-step approach was applied to the Estonian Biobank cohort of 85,050 individuals. Using both direct variants and linkage disequilibrium proxies, 10 of 14 associations (87% of testable POEs) were successfully replicated across cohorts.
Conclusions
In sum, the study presented a novel approach to infer PofO of haplotypes and detected 34 POEs, including 19 bipolar effects. By incorporating mtDNA, inference was extended to females, increasing sample size and allowing for sex-specific analysis. The analysis of imprinted regions identified multiple bipolar POEs providing compelling support for the parental conflict hypothesis. Overall, the findings reveal the broad contribution of POEs to the genetic architecture of complex traits, though with important caveats: accuracy depends on cohort-relatedness structure, and the approach cannot distinguish between genomic imprinting and parental environmental influences.