The coronavirus disease 2019 (COVID-19) pandemic raging across the world is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The receptor-binding domain of the spike proteins present on the virus envelope bind to the human angiotensin-converting enzyme 2 (ACE2), followed by fusion of the virus with the host cell membrane. The spike protein has two subunits: S1, which binds to host cells, and S2, which plays a role in membrane fusion.
.jpg)
The SARS-CoV-2's spike protein mediates viral entry into host cell. 3d illustration. Image Credit: Design Cells / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Immunizing antibodies produced by the host immune system target the RBD and disrupt the virus's binding to ACE2. However, when there are mutations in the spike protein, it may affect the efficacy of the neutralizing antibodies. Currently, there are about 930 mutations reported worldwide. A mutation from ASP614 to GLY614 has made the virus more infectious, according to reports.
Most strategies being developed that combating the virus similarly to antibodies are based on the spike protein sequence of the Wuhan reference strain. Missense mutations in the previous infectious coronaviruses like MERS and SARS-CoV have been observed to become resistant to neutralizing antibodies for the original strain. Thus, mutations in SARS-CoV-2 may also lead to strains that are resistant to the antibody treatments being developed. Hence, it is necessary to monitor mutations in the circulating SARS-CoV-2 strains to develop better therapeutics.
Simulating SARS-CoV-2 mutant binding with ACE2
Researchers from the Harbin Institute of Technology, China, modeled the complexes formed between the RBD and the human ACE2 and monoclonal antibodies and reported their findings in a paper published in the bioRxiv* preprint server.
The authors compared the complexes formed by the wild-type virus and the mutated virus and performed molecular dynamics simulations along with molecular mechanics/Poisson-Boltzmann surface area scheme. The mutations in SARS-CoV-2 were seen mainly in the open reading frame (ORF) regions, which encode for non-structural proteins, nucleocapsid protein, and spike protein. The ORF3a protein, which can change the environment inside the infected cell and make holes on the host cell membranes, has a higher mutation rate in North America and Oceania, and may allow the virus to spread better.
The authors found that mutations were not on single sites. On the spike protein, the D614G variant was the majority mutation, followed by D936Y. The N439K variant, where asparagine at the 439th site is replaced by lysine, is the most dominant in the spike protein RBD.
The molecular dynamics simulations showed more flexibility changes in the N439K variant, which could result in structural rearrangements in the SARS-CoV-2 RBD-ACE2 complex that lead to a stronger binding. Furthermore, the complex with the mutated virus forms more hydrogen bonds than the wild-type complex.
The binding energy of the N439K complex was also higher than that of the wild-type complex. This suggests that the mutant virus has a stronger association with human ACE2. The stronger binding could be because the replacement of asparagine with lysine forms a new salt bridge in the complex with human ACE2, which could increase electrostatic interactions. Apart from this interaction, the complexes are also bound by van der Waals interactions and polar solvation free energy.
N439K mutant may be resistant to some monoclonal antibodies
Although previous studies have suggested mutant versions of the virus may be less infectious, the stronger binding of human ACE2 with the N439K mutant suggests this mutant strain may be more infectious. The N439K mutation is completely included in the D614G samples, which have been observed to be more infectious than the original strain.
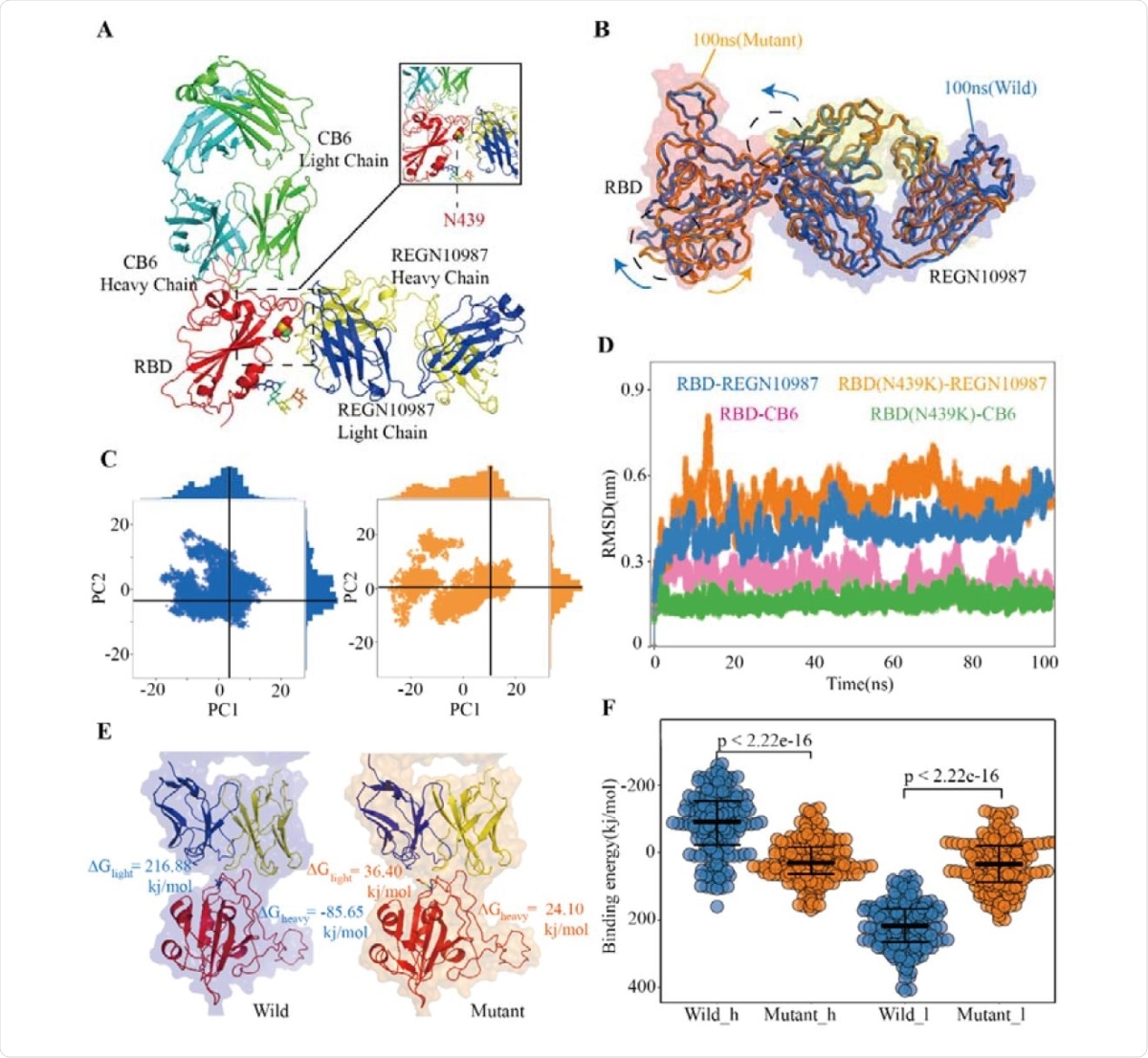
Structural and Energetic Details of Both Wild and Mutant RBD-mAbs Interactions (A) Crystal structures of RDB-CB6/REGN10987 complexes, the RBD is colored red, CB6 heavy and light chains are represented as marine and green respectively, REGN10987 heavy chain is colored yellow and light is blue, and the 439 residues are described as the sphere. (B) Characteristic dynamic fluctuations of both RBD-REGN10987 and RBD(N439K)-REGN10987 complexes. Mutant-type (100ns) and wild-type (100ns) are colored by orange and blue, respectively. (C) Dynamic conformations are projected on to the principal vectors (PC1 and PC2). Red and blue indicate mutant-type and wild-type 100ns MD trajectories respectively. (D) The RMSD of the receptor-binding motif in four complexes during the 100-ns MD simulations. (E) The binding free energies for both complexes of the mAb REGN10987 (including heavy and light chains), the color schemes are the same in Fig.4A and Fig.4C. (F) The binding free energies of 200 configurations at an interval of 100ps from the last 20ns simulations. The t-test was conducted to check the statistical significance of the difference between two systems of binding free energies. A p-value of <0.05 indicates that the difference is statistically significant (95% confidence interval). The color scheme is the same as that in Fig.4C
The authors also performed simulations of the N439K mutant of human ACE2 complexes with two neutralizing monoclonal antibodies, REGN10987 and CB6. REGN10987 binds to CR2 and CR3 regions of the RBD where N439K is located, while CB6 binds to CR1 and CR2. The analysis indicated that the N439K mutation decreased sensitivity to CB6 antibodies.
Although CB6 could neutralize the N439K mutant, the strain was quite resistant to REGN10987 antibodies. Thus, as new antiviral strategies are being developed based on the Wuhan strain, given the possible mutations in SARS-CoV-2 that could become resistant to antibodies developed for this strain, "it is necessary to consider the impact of different mutations on the effectiveness of neutralizing antibodies," write the authors.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zhou, W. et al. (2020) N439K variant in spike protein may alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. bioRxiv. https://doi.org/10.1101/2020.11.21.392407, https://www.biorxiv.org/content/10.1101/2020.11.21.392407v1
- Peer reviewed and published scientific report.
Zhou, Wenyang, Chang Xu, Pingping Wang, Meng Luo, Zhaochun Xu, Rui Cheng, Xiyun Jin, et al. 2021. “N439K Variant in Spike Protein Alter the Infection Efficiency and Antigenicity of SARS-CoV-2 Based on Molecular Dynamics Simulation.” Frontiers in Cell and Developmental Biology 9 (August). https://doi.org/10.3389/fcell.2021.697035. https://www.frontiersin.org/articles/10.3389/fcell.2021.697035/full.