The severe acute respiratory syndrome coronavirus 2 (SARS-CoV -2) has been circulating the world for almost a year, causing tens of millions of infections and well over 1.5 million deaths. This coronavirus disease 2019 (COVID-19) pandemic has been an important contributor to global economic disruption as well.
The virus has shown numerous mutations, which may affect its transmissibility or pathogenesis. Thus researchers have spent much time tracing the changes in the viral genome over time.
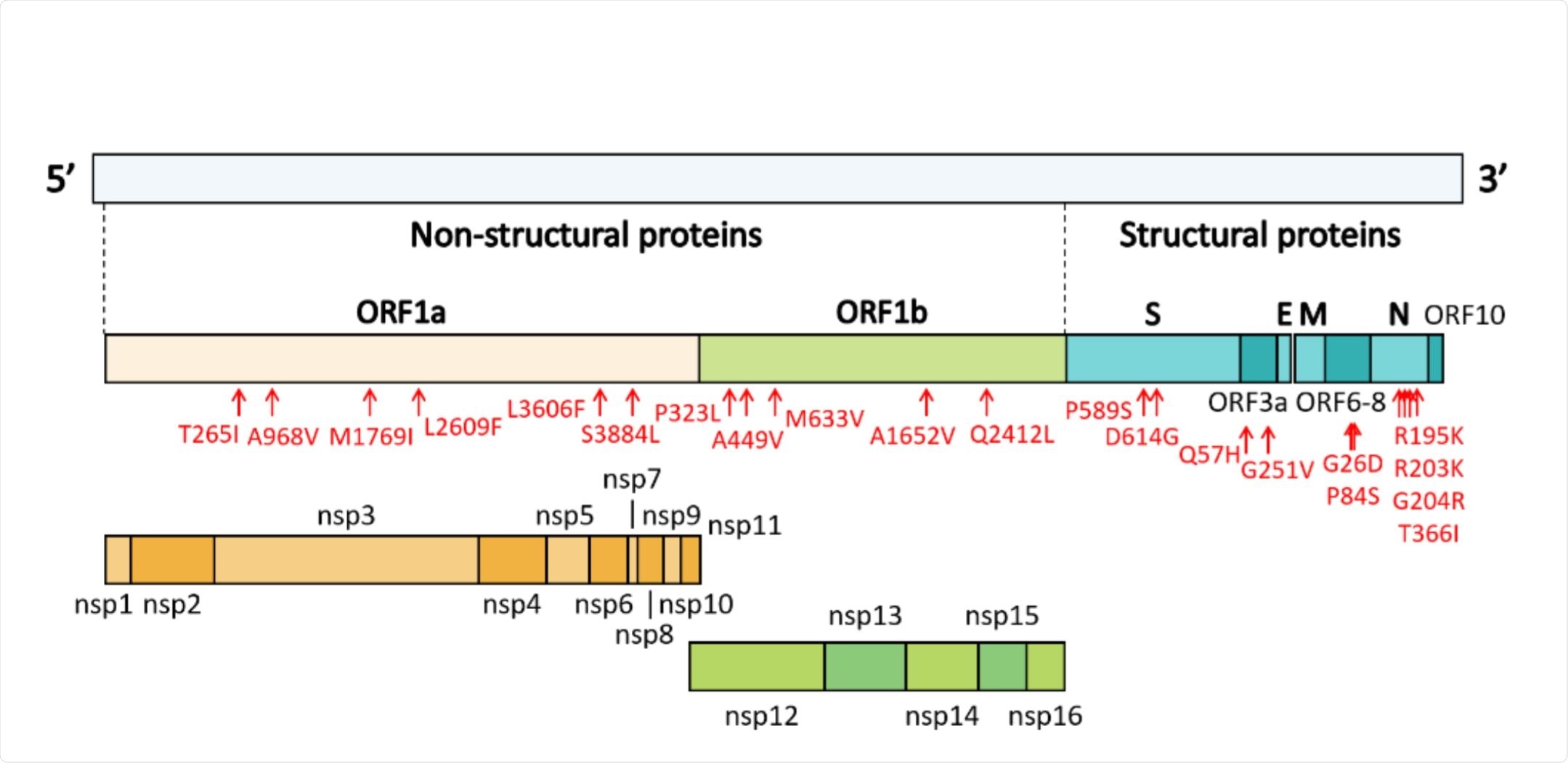
Genome organization of SARS-CoV-2. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.16.423178v1.full.pdf

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Study details
A new paper from South Korea describes the evolving SARS-CoV-2 genome in a single city, that of Gwangju, in the southwestern region of this country. The first case here was reported on February 3, 2020, following the first reported introduction of the virus into the country from China, on January 20, 2020.
The researchers, who have reported their work in a preprint on the bioRxiv* server, examined SARS-CoV-2 isolates from 10 COVID-19 patients in this city, looking for mutations of the non-synonymous kind, that is, those that caused amino acid changes.
They aligned 40 complete viral genome sequences, retrieved from the Global Initiative on Sharing All Influenza Data (GISAID; https://www.gisaid.org/ org/) database with 16 isolates from Gwangju. Using next-generation sequencing (NGS) methods, they identified point mutations in the genome.
They were thus able to trace the phylogenetic descent of the isolates and to classify the viral strains from these patients. There are three major clades, according to GISAID data, namely, S, V, and G, defined by the mutations ORF8-L84S, ORF3a ORF3a-G251V, and S -D614G, respectively.
Identifying the clades
The researchers identified three viral clades in the Gwangju sequences, namely, GH, GR, and V. Of the seven V isolates, five were from an outbreak within a religious group called the Shincheonji Church of Jesus, and two from another church. Temporally, these belonged to the early phase of the outbreak.
Nine of the isolates were from G, seven from GH, and one from GR. Of the latter, six belonged to the same cluster spread by door-to-door selling at Geumyang Building. This cluster started with one case in the building, between the second part of June and the middle of July, and spread within Gwangneuksa temple, church, and workplaces. The single GR isolate had a history of international travel.
The V clade was thus the dominant clade in Gwangju in the early phase of the outbreak in South Korea, while GH took over later in mid-July.
They explored 12 viral sequences that encoded proteins, namely, coding sequences (ORF1a, ORF1b, S, ORF3a, E, M, ORF6, ORF7 ORF7a, ORF7b, ORF8, 122 N, and ORF10) ORF10. There were 21 non-synonymous mutations and one frameshift mutation (deletion), as compared to the reference genome downloaded from the GISAID database. Leaving out the clade-defining mutations, they found 19 others, of which 11 belonged to ORF1a and ORF1b. These encode the large polyprotein that is cleaved to generate the 16 viral non-structural proteins and take up two-thirds of the whole genome.
Mutations in the viral genome
The ORF1a contained six types of mutations, most commonly the ORF1a-T265I and ORF1a-L3606F which were present in seven samples each, and in ORF1a-S3884L in six samples. Of these, ORF1a-186 L3606F occurred along with ORF3a-G251V, the V-defining mutation.
There were three other mutations within the non-structural protein (nsp) 3 within ORF1a. Since both nsp2 and 3 may contribute to differences in the infectivity of this virus, as compared to the earlier coronavirus SARS-CoV, this is a significant finding.
They also found five mutations in ORF1b, with the P323 found in nsp12 in nine sequences, A449V also being in the same protein, which encodes the RNA-dependent RNA polymerase (RdRp) enzyme, respectively. Another common mutation was Q241L, which was found in six sequences.
There were four mutations in the N protein, which is essential for the packaging of the viral RNA genome, which must happen before the virions can be released from the host cells. Of these, G204R was also found to co-occur with P323L in nsp12. These linked mutations define the GR subclade, while the GH subclade was defined by the ORF3a-Q57H mutation, respectively.
There were two mutations in ORF7 as well, in addition to a frameshift deletion of 92 nucleotides. An interesting note on this is that the ORF7a ortholog in SARS-CoV is an inhibitor of the bone marrow stromal antigen 2 (BST-2). This is a molecule that restricts the release of virus particles from the infected host cells, and which triggers the cells to enter the programmed cell death or apoptotic pathway, thus limiting the infection. Thus, this deletion could have an impact on the SARS-CoV-2 disease process and is worthy of further study.
Implications
This early study on SARS-CoV-2 isolation and mutational evolution in South Korea’s Gwangju shows that the isolates come chiefly from the V and G clades, shifting from the first to the second. Such analyses are important to understand the possible effects of mutation on viral infectivity, pathogenicity, and the way the virus interacts with the host cell.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.