Akin to all other viruses that undergo natural mutation process over time, SARS-CoV-2 made the international scientific community gravely concerned with the spread of a novel, more infectious variant of this agent that causes coronavirus disease (COVID-19).
Also known as B.1.1.7 strain, its non-synonymous mutations are specified by six mutations and two deletions in the spike glycoprotein (which is a primary vaccine target), two mutations, and an early stop codon in non-structural protein 8, as well as two mutations in the viral nucleoprotein.
Consequently, its circulation within the UK has raised public concerns as the potential for reinfection was discussed. Also, vaccine efficacy came into question due to conceivable evasion from antibody recognition. The pertinent question now is: how likely is a latter scenario?
In this study, a research group led by Dr. Winston A. Haynes from Serimmune, Inc. in Goleta (California, United States) examined both wild-type SARS-CoV-2 and the B.1.1.7 variant in order to appraise whether the mutations found in the B.1.1.7 variant make existing antibodies unable to bind to their respective viral targets.
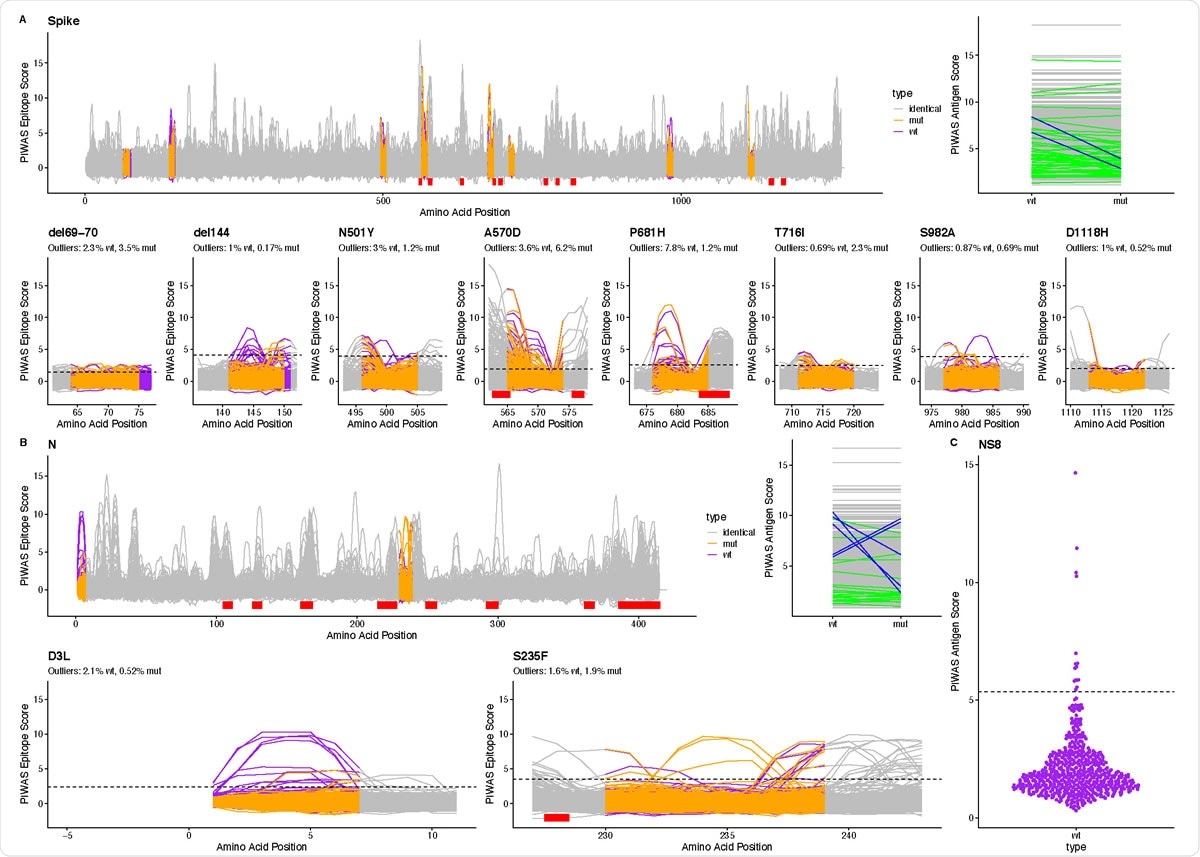
Linear epitope signal in COVID-19 patients for B.1.1.7 SARS-CoV-2. We used PIWAS analysis to examine linear epitope signal against B.1.1.7 SARS-CoV-2 in 579 COVID patients. For (A) spike glycoprotein and (B) nucleoprotein, we performed PIWAS tiling against both wild type (purple) and B.1.1.7 (orange) SARS-CoV-2. Regions outside of the variant region are tiled against the wild-type protein and are marked as identical (grey). Dominant SARSCoV- 2 epitopes identified in Haynes, Kamath, Bozekowski, et al4. are shown with red bars. PIWAS antigen score highlights individuals where the signal changed (green) or decreased by at least 3 (blue) as a result of the strain variants. For each epitope, we show the percentage of outliers (dashed line, PIWAS score > 99th percentile of the pre-pandemic control population) for the wild type and B.1.1.7 mutant. (C) We show non-structural protein 8 PIWAS antigen scores for the wild type SARS-CoV-2 strain relative to the 99th percentile in the pre-pandemic control population.
Appraising mutations and epitope signals
A high throughput, random bacterial peptide display method known as serum epitope repertoire analysis (SERA) was applied to enable the assessment of SARS-CoV-2 seropositivity and high-resolution mapping of viral epitopes across any arbitrary proteome.
More specifically, the researchers aimed to appraise whether non-synonymous mutations found in the B.1.1.7 strain can halt binding by antibodies generated during natural infection with the existing, wild-type SARS-CoV-2. For that purpose, they have performed SERA on 579 COVID-19 patients (collected before the emergence of the aforementioned mutated strain).
They have acquired a set of synthetic oligosaccharides known as 12mers for each patient, which basically represent the patient's antibody epitope binding specificities. By employing protein-based immunome wide association studies (PIWAS), the researchers tiled COVID-19 patient SERA data against both the B.1.1.7 and the wild type SARS-CoV-2 proteome in a computer model.
At the antigen level, the obtained peak wild-type and peak mutant PIWAS values were compared to pinpoint individuals with decreased epitope signal at the antigen level. In a nutshell, they have compared PIWAS antigen scores for the same proteins and variants.
Limited changes in comparison to the original strain
This study showed that the mutations reduced signal substantially in only 0.5 percent of the population at the antigen level. Albeit some epitope mutations can diminish measured signal in up to 6 percent of the population, these are not the dominant epitopes for their antigens.
Changes conferred by mutations resulted in only two individuals (i.e., 0.3 percent of the population) having a considerable reduction in PIWAS antigen scores, reflecting the peak epitope signal across the entire antigen. Basically, COVID-19 patients had limited changes in epitope signal on the B.1.1.7 strain when compared to the original strain.
And although the early stop codon in accessory or non-structural protein 8 of the SARS-CoV-2, an ORF8 gene product – resulted in the most decreased signals, antibodies to non-structural protein 8 are not likely to play an important role in a protective response.
No loss of vaccine efficacy
"Our data suggest that the mutations seen in the B.1.1.7 strain of SARS-CoV-2 would not result in loss of dominant antibody responses to linear spike glycoprotein and nucleoprotein epitopes in the vast majority of our cohort's COVID patients", say study authors in this medRxiv paper.
Since PIWAS can detect linear epitope signals, further assays are necessary to evaluate structural evasion of the immune response. In particular, the infamous mutation P681H is found just upstream of the furin cleavage site, which is actually a hotspot for epitope signal.
However, as mRNA vaccines include intact spike sequences (like those circulating in naturally infected individuals), and naturally infected indeed show a durable immune response, there is currently no evidence to suggest that the existing vaccines will not be effective against B.1.1.7. In the meantime, the situation has to be continuously monitored.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.