As the coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), refuses to subside, the search for effective antivirals and therapeutics continues without respite. An encouraging paper published in the journal Nature Communications on January 12, 2021, describes a promising new therapeutic antibody that potently neutralizes the virus.
Globally, the D614G-mutation-bearing viral variant has become dominant in almost every place. Many scientists consider this strain to be more transmissible because of its increased efficiency of viral entry. The RBD is thus a prime target for antibodies specific to the SARS-CoV-2 virus, but produces little or no antibody-dependent enhancement (ADE) of disease.
The current paper deals with the monoclonal antibody (mAb) CT-P59, which strongly binds the RBD, offers steric hindrance to the binding of the RBD to the ACE2 receptor, and reduces symptoms of the disease in vivo, besides the in vitro efficacy it demonstrates.
Isolation of CT-P59
CT-P59 is a mAb in the form of fully human immunoglobulin G (IgG). It was shown, by a plaque reduction neutralization test (PRNT), to reduce SARS-CoV-2 replication substantially, with a low half-maximal inhibitory concentration (IC50) of 8.4 ng/mL. This was so with the ancestral as well as the D614G variant, with the IC50 for the latter being less, at 5.7 ng/mL.
The use of biolayer interferometry (BLI) showed that this antibody fully suppressed RBD-ACE2 binding. CT-P59 bound to RBD mutants as well, maintaining complete inhibition of binding. This suggests that mutational escape has not yet occurred with naturally occurring RBD variants, since its binding to the RBD does not involve the mutated sites.
The antibody was found to be highly specific for SARS-CoV-2 relative to other coronaviruses. Finally, the use of surface plasmon resonance analysis showed the high affinity of CT-P59 for the virus.
Structural study of neutralization by CT-P59
The researchers carried out X-ray crystallography at 2.7 Å resolution. This showed that the antibody binds the ACE2-interacting surface, called the receptor-binding motif (RBM), within the SARS-CoV-2 RBD. While this is common among such mAbs, the angle of association between the RBD and the CT-P59 is different from that of most other antibodies. CT-P59 binds to the RBD without steric hindrance only when it is in the ‘up’ conformation.
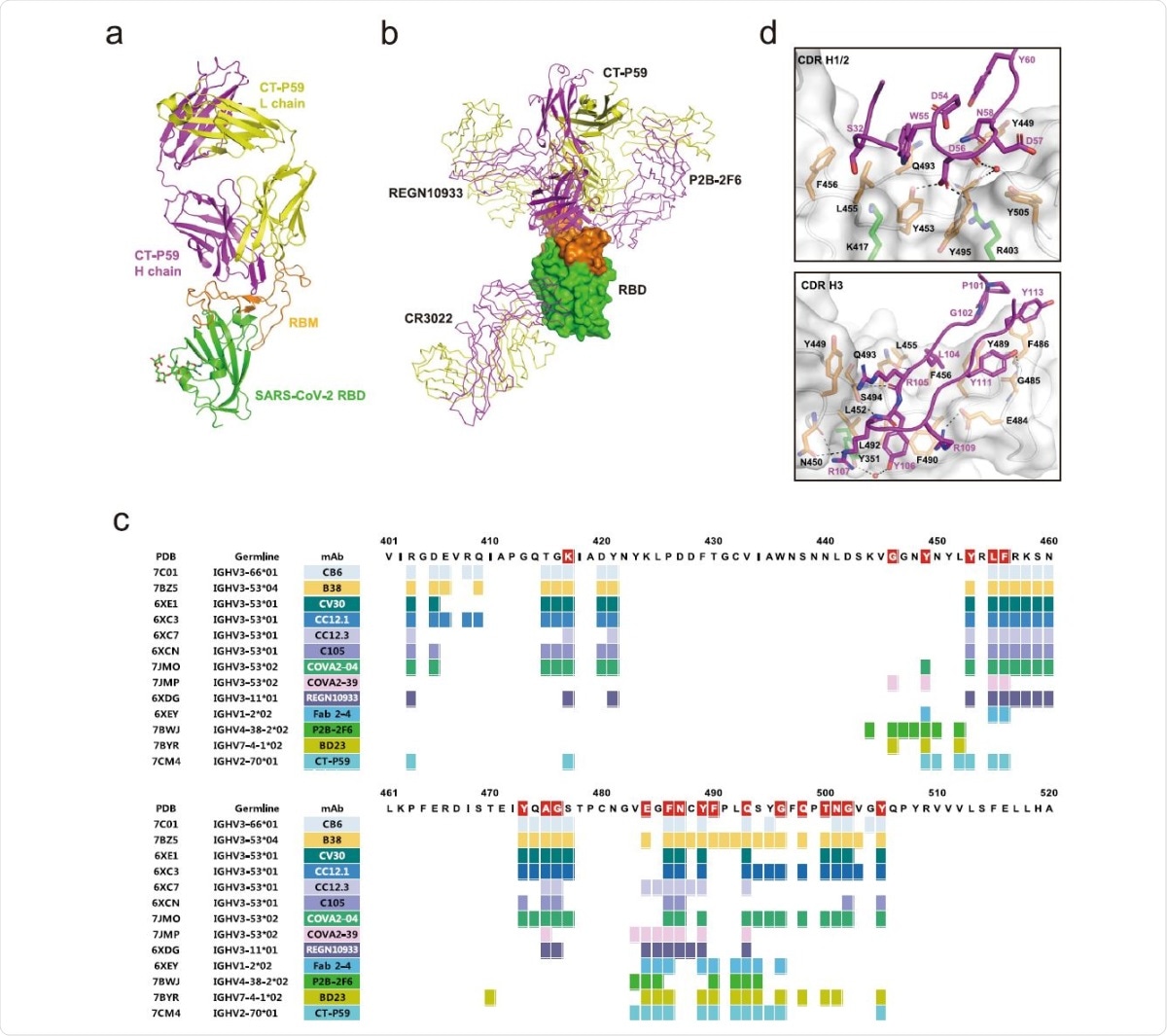
The overall structure of the CT-P59 Fab/SARS-CoV-2 RBD complex. The RBD domain is green for the core subdomain, and orange for RBM. The heavy and light chains of CT-P59 are magenta and yellow, respectively. b Superposition of the neutralizing antibodies in complex with RBD. RBD is shown as a surface model. CT-P59 is shown as a cartoon, and the other antibodies (CR3022: PDB 6XC3, PB2-2F6: PDB 7BWJ, REGN10933: PDB 6XDG) are shown as a ribbon model. The heavy and light chains of Fab are magenta and yellow, respectively. c Assignment of the epitope residues for RBD-targeting neutralizing antibodies with a distance cutoff of 4.5 Å. RBD residues interacting with ACE2 are highlighted in red. d The detailed interactions between the RBD and CDR loops of CT-P59. The interfaces between RBD and CDR H1/2 or H3 are shown in the top and bottom panels, respectively. The RBD domain is shown as a surface model with a semi-transparent representation. The CDR loops and interacting residues on the interfaces are shown in ribbons and sticks, respectively. The residues are colored as in a. Dashed lines indicate hydrogen bonds. Water molecules are shown as red spheres.
Most neutralizing antibodies have a common immunoglobulin heavy-chain variable region genes (IGHV) 3 germline origin, whereas CT-P59 is based on IGHV2. In fact, this is the first time the high-resolution structure of any IGHV2 germline RBD-neutralizing antibody has been reported. The angle of association of CT-P59 is similar to that of another IGHV3 germline antibody, COVA2-39, which is different from other IGHV3 antibodies.
Most of the protein-protein interactions (PPIs) between the RBD and receptor involve the heavy chains, at all three complementarity determining regions (CDRs), with 16 and 19 residues of the antibody and the RBD, respectively. There is an 18-residue β-hairpin structure of the CDR H3 that forms eight hydrogen bonds as well as many hydrophobic interactions with aromatic amino acids central to the ACE2 binding surface. This loop is thus key to the strong PPIs mediating this association. The light chains show only weak contacts at three points.
The RBD- CT-P59 interaction may produce a local change in the conformation of the β5–β6 loop region, with the heavy chain overlapping the ACE2-binding site completely, and the light chain showing partial overlap. Thus, this antibody completely blocks the binding surface of the receptor.
In vivo efficacy in animal studies
The researchers used ferrets, golden Syrian hamsters, and rhesus monkeys to evaluate the in vivo efficacy of neutralization. With ferrets, the antibody reduced viral RNA and infectious virus titer in the nasal wash at 3 days post-infection (dpi), with undetectable infectious virus by 6 dpi. The lung tissue also showed low infectious virus titers at 3 dpi, which became undetectable by 7 dpi. Clinical symptoms, and lung pathology, were also reduced in keeping with these parameters.
In comparison, remdesivir, which is a US FDA approved drug, failed to achieve undetectable infectious virus in nasal wash by 6 dpi, indicating that this drug takes longer to clear the virus than CT-P59.
At the same dose of 30 mg/kg, CT-P59 achieved infectious virus clearance from the lungs within 48 hours, indicating that it fully suppressed viral replication. With rhesus monkeys, both CT-P59-treated animals and controls remained healthy. However, in the latter group, the viral load peaked by 2 dpi and then went down by 6 dpi. The CT-P59-treated group showed a rapid decline in viral titers, and viral clearance from the upper respiratory tract, at 2 dpi, at 45 mg/kg. Viral RNA was found in lung tissue, but no infectious virus in either group.
In vitro studies
The use of CT-P59 in cell cultures along with wildtype SARS-CoV-2 failed to show any ADE-related effects mediated by the Fc receptors.
Conclusion
The researchers sum up: “CT-P59 mAb, along with small molecule drugs such as remdesivir and dexamethasone, may thus help curb pandemic as a therapeutic or preventative intervention for COVID-19.” It has already passed phase I clinical trials and phase II trials are ongoing in several countries.