Several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern are characterized by mutations in the receptor-binding domain and more significant spike protein that enhance virulence by increasing the affinity of this region to the target ACE2 receptor found on the surface of cells, and could potentially better evade vaccine-generated antibodies that are specific to the wildtype spike protein. The genome of SARS-CoV-2 has been subject to continuous scrutiny since the early stages of the pandemic, and currently, nearly a million sequences have been publicly shared through various online databanks. This has allowed a previously unheard-of degree of real-time analysis of the changing nature of the genome, which has been exploited in a paper recently uploaded to the preprint server bioRxiv* by Giacomo et al. (March 29th, 2021) to identify a novel spike protein mutation that has risen in frequency among the population since early 2021.
Frequent novel SARS-CoV-2 mutations
The group accessed 827,572 SARS-CoV-2 genomic sequences publically available from the global science initiative online databank as of March 26th, 2021, and compared them to the reference SARS-CoV-2 wildtype genome, uncovering nearly 17 million mutation events. Upon translating these sequences into three-dimensional protein structures, the group identified novel mutation T478K in the spike protein ACE2 interaction complex, which existed in 4,214 COVID-19 patients.
Variant B.1.1.222 was first detected in Mexico in April 2020, and mutation T478K is present in around 65% of occurrences of this strain. Of the 4,214 samples discovered to bear this mutation by the group, 86% were of the B.1.1.222 variant. The remainder were of various distinct lineages, suggesting that this mutation has occurred in multiple distinct mutation events. This mutation was present in 38.1% of all cases in Mexico and 1.3% in the USA and was also detected infrequently in some European countries.
The frequency of the T478K mutation has risen exponentially since early 2021, similarly to another mutation in the spike protein: D614G, which the group also tracked over time. D614G was reported to emerge shortly before T478K and began to rise quickly in frequency in late 2020. Currently, around 90% of SARS-CoV-2 genomes bear the D614G mutation, exhibiting the rapid rise to prominence of this mutation, which reportedly enhances virulence by promoting entry into the host cell. Currently, only around 2% of the total SARS-CoV-2 population bears the T478K mutation, according to this work, though if the mutation enhances virulence similarly then it may correspondingly rise to dominance.
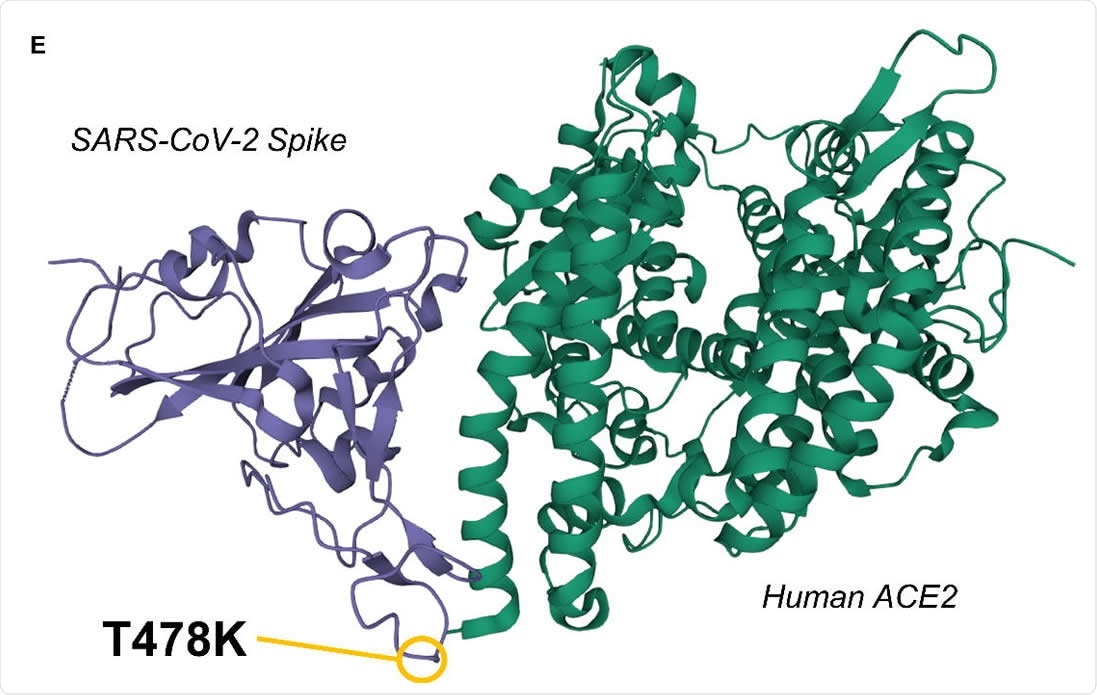
3D representation of the SARS-CoV-2 Spike / Human ACE2 interacting complex

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The T478K mutation
The T478K mutation constitutes the exchange of the non-charged amino acid threonine with the positively charged lysine at position 478 and roughly encompasses amino acids 350 to 550 of the SARS-CoV-2 spike protein. The exchange of amino acids at this position facilitates the presentation of a different electrostatic surface that can be further altered by co-occurring mutations, potentially interacting with receptors, antibodies, and drugs more strongly or weakly. Three other spike mutations were found to occur in parallel with T478K frequently: D614G, which represents one of the founding mutations of the B lineage and thus had co-occurrence of 99.8%; P681H, with 95.2% co-occurrence; and T732A, with 91.4% co-occurrence. How these amino acids interact to enhance the virulence of SARS-CoV-2 is currently unknown. However, the authors highlight another group's results, where T478K and T478R mutations were observed to develop in SARS-CoV-2 treated with weakly neutralizing antibodies, suggesting that this mutation could aid in evading the immune response.
There is not yet any clinical indication that T478K enhances virulence, however, the positioning of the mutant amino acid at the tip of the interaction point with ACE2 in association with the rapid rise in frequency recently observed in Mexico and the southern USA may indicate that this is the case. Mutations in this sensitive region of the spike protein often reduce affinity, and thus the fact that the variant is multiplying in the human population suggests survival validity. This work underscores the importance of monitoring the occurrence and spread of SARS-CoV-2 mutants to better advise public policy and vaccine development.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Preliminary report on SARS-CoV-2 Spike mutation T478K Simone Di Giacomo, Daniele Mercatelli, Amir Rakhimov, Federico M Giorgi, bioRxiv, 2021.03.28.437369; doi: https://doi.org/10.1101/2021.03.28.437369, https://www.biorxiv.org/content/10.1101/2021.03.28.437369v1
- Peer reviewed and published scientific report.
Di Giacomo, Simone, Daniele Mercatelli, Amir Rakhimov, and Federico M. Giorgi. 2021. “Preliminary Report on Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) Spike Mutation T478K.” Journal of Medical Virology, May. https://doi.org/10.1002/jmv.27062. https://onlinelibrary.wiley.com/doi/10.1002/jmv.27062.