Membrane proteins are heavily involved in a variety of cellular processes such as signal transduction, membrane translocation, cell adhesion, and many more, and therefore are of great interest within the field of drug design.
The structure of relevant membrane proteins is sometimes mimicked during research using detergent micelles, which are capable of entrapping and extracting the membrane protein within a lipid layer that somewhat emulates the conditions found within the lipid bilayer of the cell membrane.
However, detergents can interfere with binding interactions that the protein may have otherwise made in their absence and can influence tertiary protein structure.
Similarly, phospholipid liposomes and other types of lipid bilayer have been employed to mimic the natural conditions of membrane proteins. However, these techniques often result in heterogeneously sized particles that are not compatible with many microscopy techniques.
Phospholipid nanodiscs are constructed from a lipid bilayer surrounded by amphipathic proteins that corral the structure into a disc shape, producing an environment more representative of that found naturally in the cell membrane.
Unfortunately, as with liposomes, nanodiscs often suffer from non-homogenous sizing and protein loading, particularly when made in larger sizes above 16 nm in diameter.
In a paper recently uploaded to the journal Frontiers in Biotechnology by Padmanabha Das et al. (June 12th, 2020), a method of producing significantly larger homogenous nanodiscs up to 90 nm in diameter is described, and the advantages and applications of such nanodiscs are explored: better replicating the actual curvature and membrane symmetry of a real cell membrane, and allowing much larger or multiple membrane proteins to be loaded simultaneously.
Nanodiscs as cell membrane mimics
Smaller nanodiscs have been produced by shortening or lengthening the amphipathic membrane structural proteins that encircle the lipid bilayer. The group utilized folded DNA in a barrel-like structure to produce larger nanodiscs, which are subsequently flooded with lipids and the membrane protein of interest.
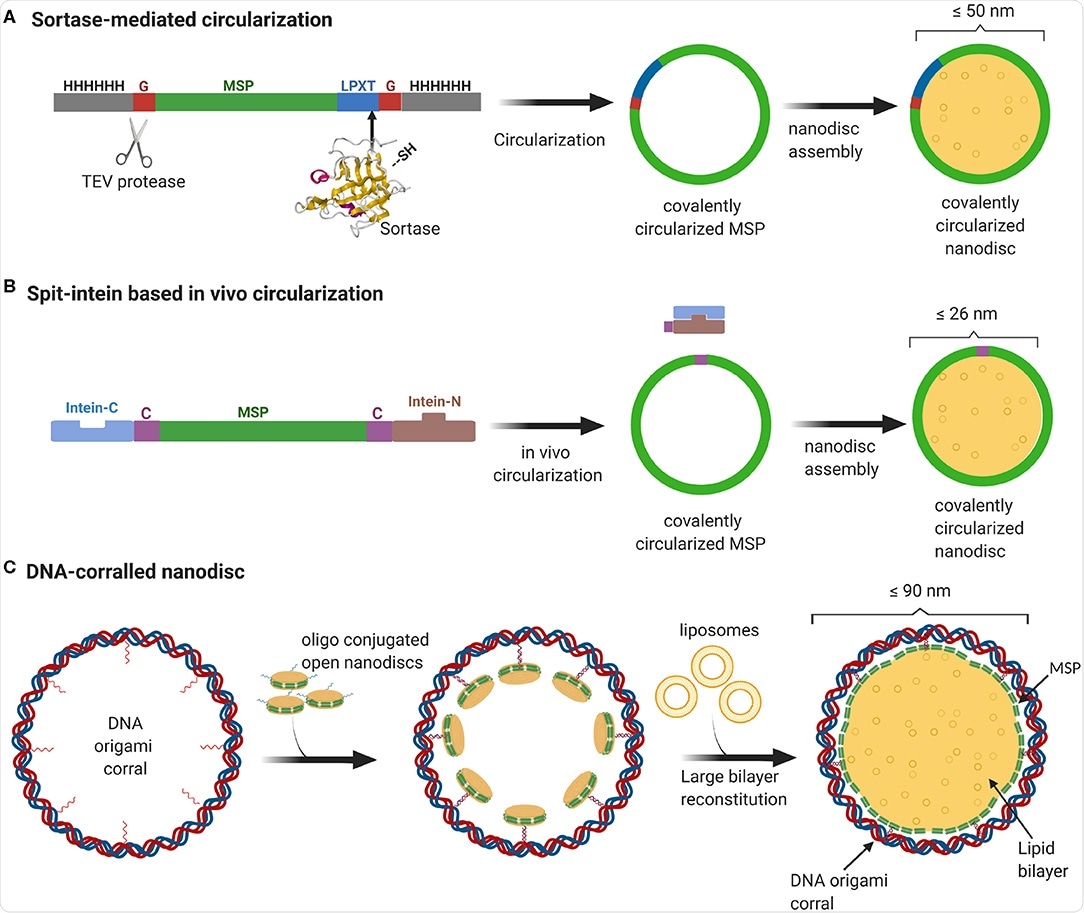
Overview of currently available methods to produce stable and homogeneous large nanodiscs. (A) Sortase-based MSP circularization strategy. Sortase cleaves between the Gly and Thr of the LPXTG motif at the C-terminus of MSP and catalyzes the formation of an amide bond with the N-terminal Gly resulting in a circular MSP which can be used to assemble nanodiscs of sizes ranging from 9 to 50 nm. (B) Split intein-based strategy for circularization of MSP in vivo using Npu DnaE split-intein. MSP fusion results in nanodiscs as big as 26 nm. (C) Outline of DNA-corralled nanodisc protocol. Small nanodiscs functionalized with oligos bind to specified sites on the DNA origami resulting in a small nanodisc-decorated barrel. Addition of detergents and lipids, followed by removal of detergents using dialysis, leads to reconstitution of large nanodiscs within DNA barrel.
The way in which a virus enters a cell varies based on the specific virus or host cell and has been the subject of great scrutiny historically and in particular more recently with the advent of the COVID-19 pandemic.
Cryo-electron microscopy allows the process to be visualized and has previously been employed in combination with liposomes to describe how, for example, human rhinovirus 2 particles cross the cell membrane. However, issues relating to homogeneity in size and loading limit the relevance of this platform.
Large nanodiscs that can support large protein complexes better mimic the natural cell membrane and are more suited to cryo-EM studies, potentially providing the ideal structure upon which these studies can be performed.
Nanodiscs prepared in this way also have the advantage of potentially being produced asymmetrically, with differing “inner” and “outer” membranes, as observed naturally within the eukaryotic plasma membrane, where sphingomyelin and phosphatidylcholine are actively segregated to the outer membrane and phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol to the inner.
In conclusion, DNA origami has enabled the construction of large nanodiscs that strongly mimic the lipid bilayer found in cells, capable of supporting large or high numbers of protein complexes in an in situ-like environment.
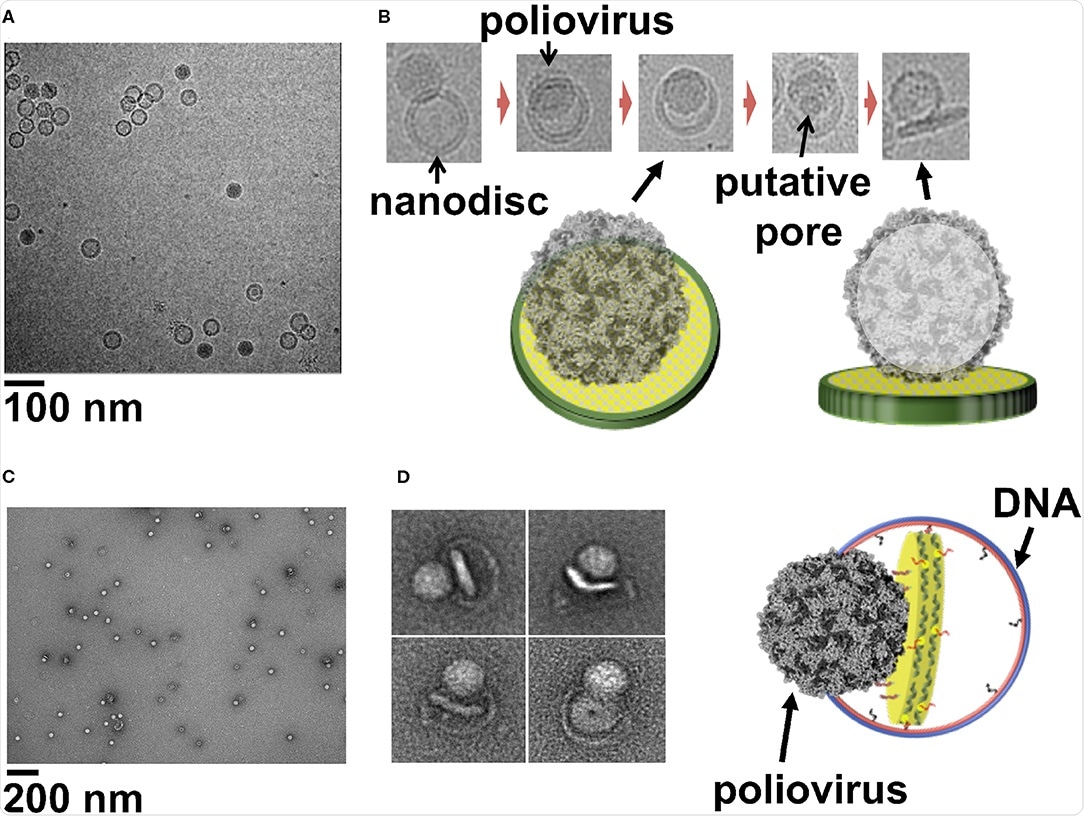
Cryo-EM image of a field view of poliovirus interacting with 50 nm cND containing CD155 receptor. (B) Cryo-EM images of individual viral particle tethered to nanodisc containing CD155. (C) Negative-stain TEM images of poliovirus interacting with 60 nm DCND containing CD155 receptor. (D) TEM images of individual viral particles interacting with DCND containing CD155. Some of the nanodiscs were partially released from their DNA scaffolds after binding the virus. The images show bending of the bilayer and creation of a pore in nanodisc by the poliovirus. A Cartoon representation of a nanodisc partially released from its DNA scaffold upon binding to the virus (right). This figure is adapted and used with permission from references (Nasr et al., 2017; Zhao et al., 2018).
This technology has a wide range of potential uses in biology, particularly concerning studying the method of cell entry employed by a virus, where the mechanism of membrane penetration, stages of virus particle disassembly, and any associated conformational change or surface receptor involvement can be established.
Journal reference:
- Padmanabha Das Krishna M., Shih William M., Wagner Gerhard, Nasr Mahmoud L., Large Nanodiscs: A Potential Game Changer in Structural Biology of Membrane Protein Complexes and Virus Entry, Frontiers in Bioengineering and Biotechnology, DOI=10.3389/fbioe.2020.00539, https://www.frontiersin.org/article/10.3389/fbioe.2020.00539