A majority of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) research focuses on the spike protein as it is crucial for viral entry and subsequent infection. Moreover, special attention has been given to the receptor-binding domain of the spike protein. However, another region called the N-terminal domain is also involved in viral entry into the host cell through sialic-acid receptor binding.
The N-terminal domain has a fold that can bind sugar, and new research led by Jonathan Lees from Oxford Brookes University found the sugar-binding pockets near the N-terminal domain help to increase viral infectivity. Furthermore, the findings show that the SARS-CoV-2 sugar-binding pockets are evolving with added loops observed in the pockets of new variants.
SARS-CoV-2 variants of concerns Gamma, Delta Plus, and Omicron have mutations in the N-terminal domain located near sugar-binding pockets and are associated with increased transmission. Understanding the structure of sugar-binding pockets on the N-terminal domain could help create new antiviral drugs.
The study “Insertions in the SARS-CoV-2 Spike N-Terminal Domain May Aid COVID-19 Transmission” was recently posted to the bioRxiv* preprint server.
 Study: Insertions in the SARS-CoV-2 Spike N-Terminal Domain May Aid COVID-19 Transmission. Image Credit: NIAID
Study: Insertions in the SARS-CoV-2 Spike N-Terminal Domain May Aid COVID-19 Transmission. Image Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Pockets in the N-terminal domain have strong binding to sugar
The researchers analyzed four binding pockets in the SARS-CoV-2 N-terminal domain along with the N-terminal domain of other coronaviruses.
Findings showed that the second and third pockets had greater binding towards sialic acid than the first sugar-binding pocket.
The binding strength of the second pocket differed across all studied coronaviruses. However, the third pocket retained strong binding to sialic acid.
Additionally, insertions in the N-terminal domain contributed to more loops that extended the first pocket. The end result was increased contact and binding with sialic acid. Other regions in the N-terminal domain near the sugar-binding pockets also enhanced sugar-binding interactions.
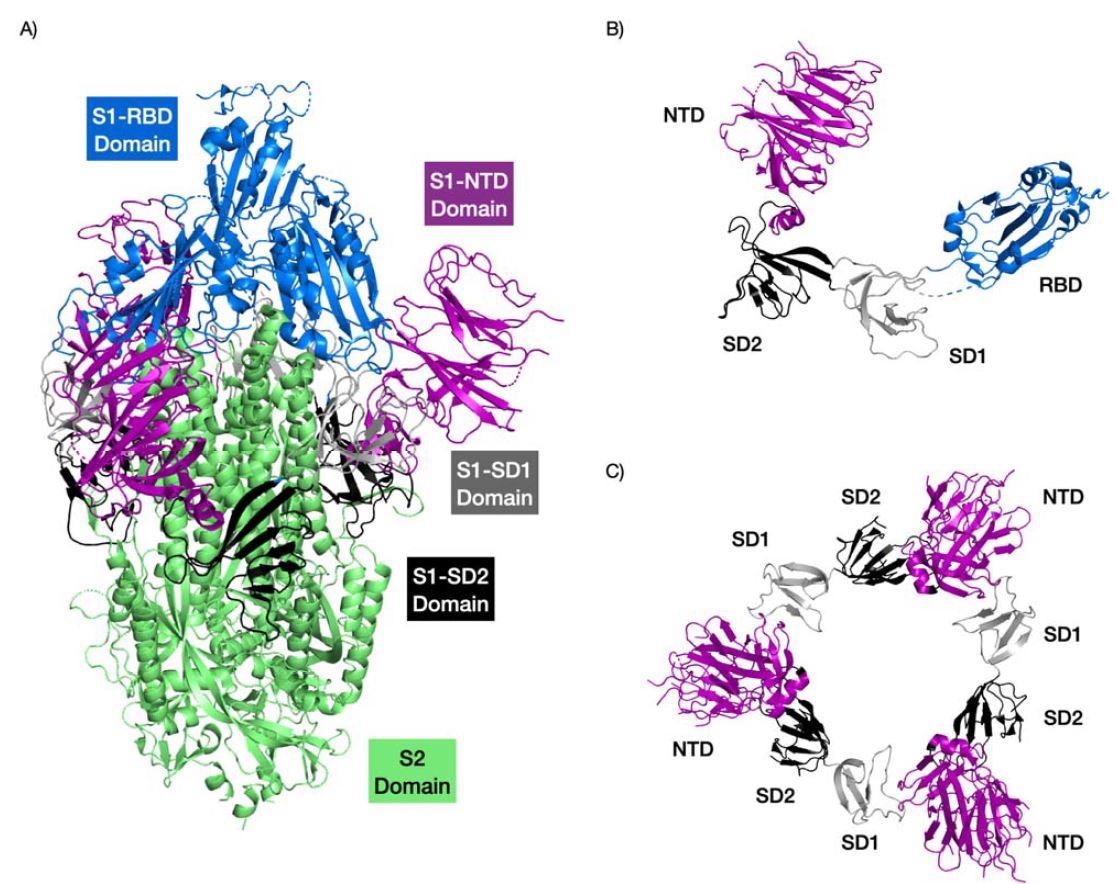 Structure of the SARS-CoV2 Spike protein highlighting the different domains. The RB domain is colored in blue, the NTD domain in purple, the SD1 domain in grey, and the SD1 domain black. The trimer complex is shown in (A). The S1 region of a Spike protein monomer is shown (B). The interactions of the NTDs with SD1 and SD2 domains of the Spike protein trimer are shown in (C). PDB structure 6VSB.
Structure of the SARS-CoV2 Spike protein highlighting the different domains. The RB domain is colored in blue, the NTD domain in purple, the SD1 domain in grey, and the SD1 domain black. The trimer complex is shown in (A). The S1 region of a Spike protein monomer is shown (B). The interactions of the NTDs with SD1 and SD2 domains of the Spike protein trimer are shown in (C). PDB structure 6VSB.
Evolution of binding pockets in newer coronavirus strains
Pockets 2 and 3 were more diverse in their structure. Additionally, the emergence of the IR2 indel region — an area with a sugar-binding motif for sialic acids — in the N-terminal domain is able to interact with pockets 2 and 3 because of the added loop. The findings suggest the SARS-CoV-2 virus is evolving in these areas to improve sialic acid-binding and increase infectivity.
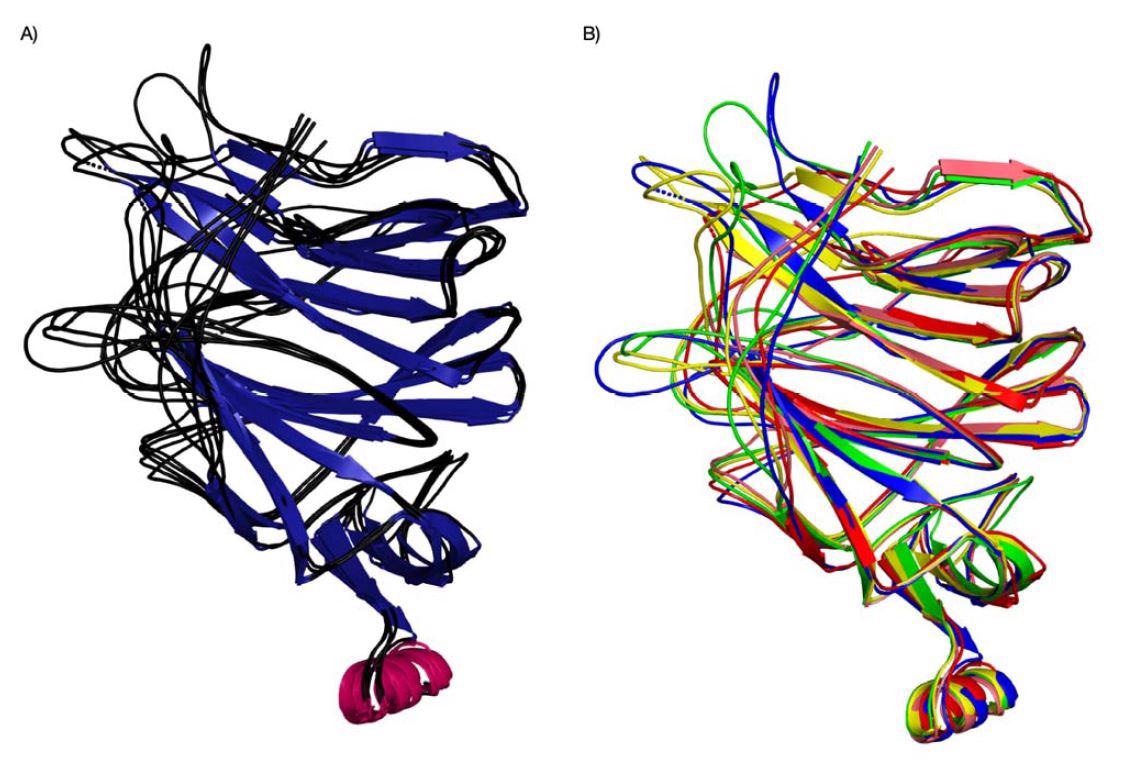 Structure superposition of Cov NTDs (SARS-CoV, PDB ID 6ACC, Pangolin CoV G PDB ID 7CN8, Pangolin-CoV-GD, PDB-ID 7BBH, Bat-Cov-RaTG13, PDB ID: 7CN4, SARS-Co 2, PDB ID 7C2L). The structures are coloured based on their secondary structure components ( and proteins (different species/variants) (B).
Structure superposition of Cov NTDs (SARS-CoV, PDB ID 6ACC, Pangolin CoV G PDB ID 7CN8, Pangolin-CoV-GD, PDB-ID 7BBH, Bat-Cov-RaTG13, PDB ID: 7CN4, SARS-Co 2, PDB ID 7C2L). The structures are coloured based on their secondary structure components ( and proteins (different species/variants) (B).
The researchers hypothesize that the SARS-CoV-2 virus may be evolving specifically in these areas as a response to specific sugar modifications in human cells. Indeed, binding energy studies showed that some SARS-CoV-2 variants of concern had increased binding to sugars in pocket 3.
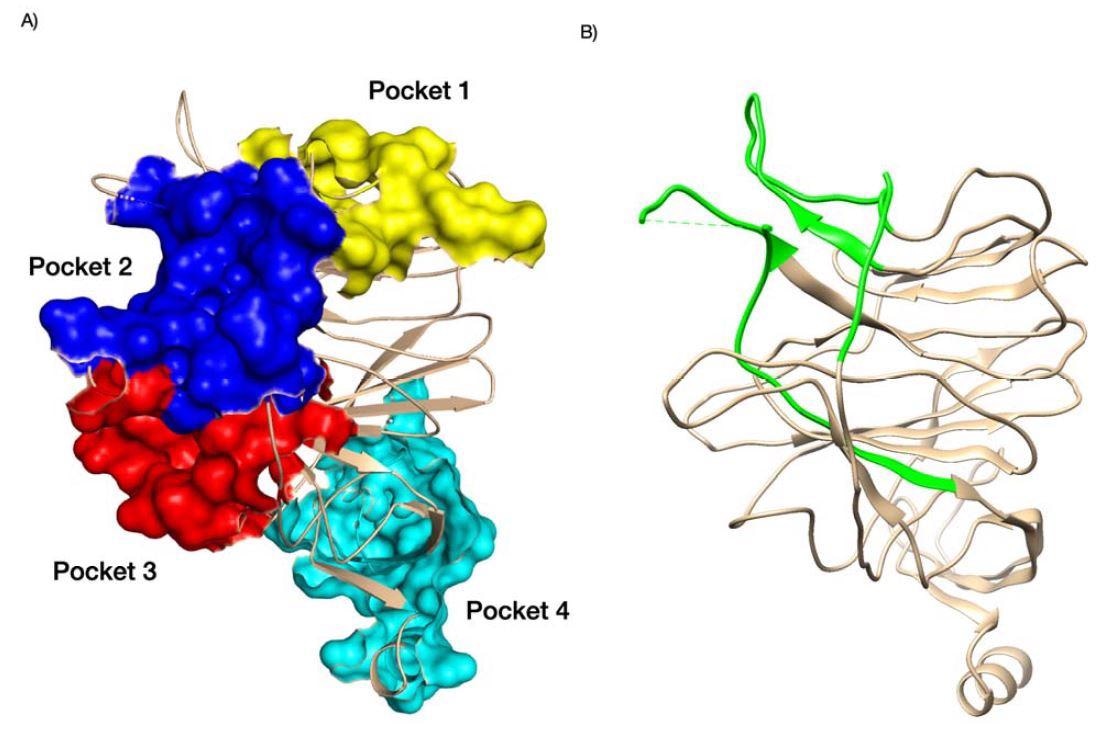 Pockets identified in the NTD domain of SARS-CoV-2 (A) and antigenic NTD supersite SARS-CoV-2 are shown in green (B). For (A), we colored pocket 1 in yellow, pocket 2 in blue, pock 3 in red, and pocket 4 in cyan. PDB structures 7C2L. Supporting references: Behloul et al., 202 Fantini et al., 2020; Baker et al., 2021; Di Gaetano et al., 2021; McCallum et al., 2021.
Pockets identified in the NTD domain of SARS-CoV-2 (A) and antigenic NTD supersite SARS-CoV-2 are shown in green (B). For (A), we colored pocket 1 in yellow, pocket 2 in blue, pock 3 in red, and pocket 4 in cyan. PDB structures 7C2L. Supporting references: Behloul et al., 202 Fantini et al., 2020; Baker et al., 2021; Di Gaetano et al., 2021; McCallum et al., 2021.
Binding affinity to sugar differ across coronaviruses and SARS-CoV-2 variants of concern
The researchers conducted a computational analysis to measure the binding energy of coronaviruses to sialic acid.
Their results found stronger binding activity in the SARS-CoV-2 N-terminal domain compared to SARS-CoV. Specifically, binding was strongest in binding pockets 1 to 3.
Compared to SARS-CoV and the original SARS-CoV-2 strain identified in Wuhan, China, the Kappa variant with the E154K mutation had stronger binding in pocket 1. In addition, the Delta, Iota, and Mu variants also had stronger binding in pocket 3 when they had the T95I mutation.
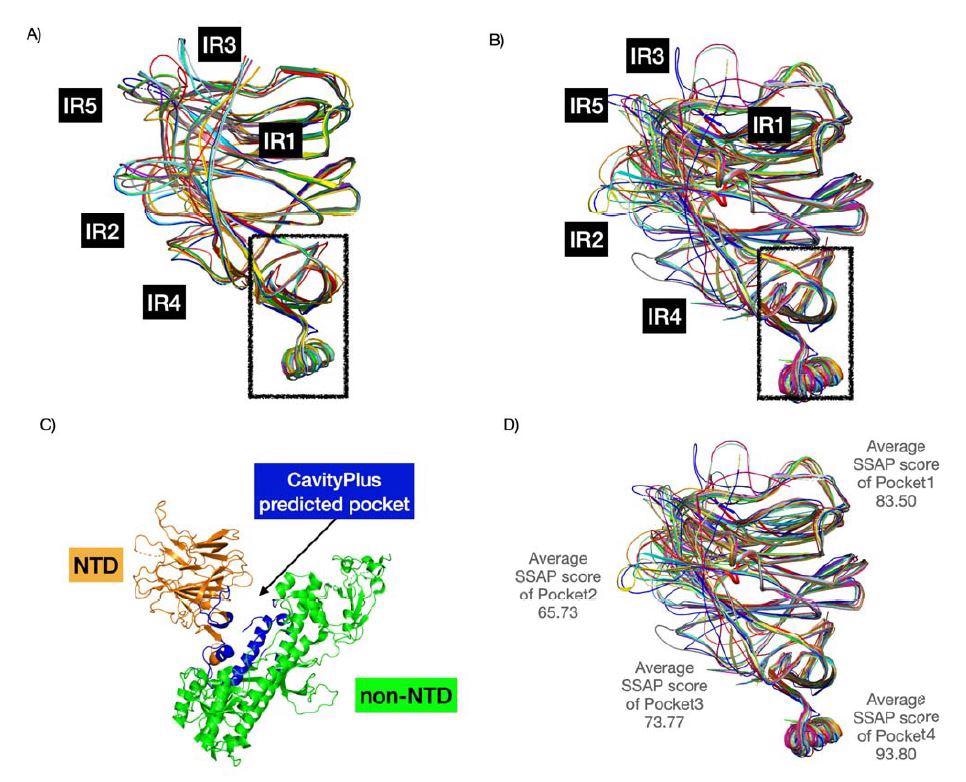 Structure superposition of CoV NTDs. A) comprises the 16 BCoV NTDs in the multip sequence alignment (see Figure 5). B) constitutes 24 BCoV NTDs from the CATH family. For and B), we used known structures and structural models (built using AlphaFold2). We found a high conserved pocket (in the box) (score of 1838 - highly positive DrugScore constitutes hi druggability) that could be a good pan coronavirus drug target. C) demonstrates this pocket prediction by CavityPlus in blue. D) We computed the structural conservation of pockets by calculating the average SSAP score.
Structure superposition of CoV NTDs. A) comprises the 16 BCoV NTDs in the multip sequence alignment (see Figure 5). B) constitutes 24 BCoV NTDs from the CATH family. For and B), we used known structures and structural models (built using AlphaFold2). We found a high conserved pocket (in the box) (score of 1838 - highly positive DrugScore constitutes hi druggability) that could be a good pan coronavirus drug target. C) demonstrates this pocket prediction by CavityPlus in blue. D) We computed the structural conservation of pockets by calculating the average SSAP score.
T95I mutations are one of the newer mutations found in the Delta plus variant.
An interesting observation to the researchers is that the distance between mutations to the sugar-binding pockets can influence binding to other regions near the pocket even if they are not directly binding to sialic acid.
The Gamma, Omicron, and Delta Plus variants have high binding energy in the N-terminal domain binding pockets 1 and 3. This may be linked to the increased level of transmission observed with these variants of concern.
“We propose continuous monitoring of NTD mutations and indels in the context of newly emerging variants and their impacts on sugar-binding,” wrote the research team. “For example, the Omicron variant has a rather unique 3 amino acid insertion at position 214 which lies near to pocket 3.”
The high conservation and druggable nature of pocket 1 make it a suitable target for the development of drugs that would reduce or block binding to sialic acid. The researchers note that targeting galectins and the sugar-binding pockets could help prevent infectivity into the host cell and influence the immune response towards the virus.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.