The coronavirus disease 2019 (COVID-19) pandemic has caused over 5.44 million deaths since its emergence in late 2019 in China. More than 9.2 billion doses of COVID-19 vaccines have been administered to date; however, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to infect both vaccinated and unvaccinated individuals, particularly in countries with low vaccination rates, due to the unavailability of vaccines or vaccine hesitancy.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Several coronaviruses (CoVs) are known to infect humans, but only three zoonotic CoVs, including the SARS-CoV of 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) of 2012, and SARS-CoV-2, are known to cause lethal infections. These three CoVs belong to the same beta coronavirus group but have different lineages and all cause acute lung injury and hypoxemic respiratory failure.
All CoVs share similar genomic structures, replication patterns, portal of entry, and show tropism for respiratory epithelia. However, these CoVs also show differences in host-virus interactions by expressing lineage-specific distinct accessory proteins.
Phylogenetic tree of betacoronaviruses and their lineages. Viruses examined in this study are shown in red font.
In a recent study published on the preprint server bioRxiv*, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was found to differ from other viruses in the beta coronavirus family in the activation of a key pathway associated with endoplasmic reticulum (ER) stress.
About the study
In the present study, researchers investigated whether inositol-requiring enzyme 1α (IRE1α)/ X-box binding protein 1 (XBP1) of the unfolded protein response (UPR) is required in human lung epithelial cell lines A549 and Calu-3, as well as in induced pluripotent stem cell (iPSC)-derived type II alveolar (iAT2) cells, upon infection with four beta CoVs. The CoVs studied in this study include human coronavirus OC43 (HCoV-OC43), murine coronavirus (MHV), MERS-CoV, and SARS-CoV-2.
About one-third of eukaryotic proteins are synthesized through co-translational translocation into the ER lumen, including the secretory and membranous proteins. Similarly, viral membrane-bound proteins are synthesized and processed in the ER.
Further, It is known that CoV replication induces the ER stress, which is detected by three transmembrane proteins, including activating transcription factor 6 (ATF6), PKR-like ER kinase (PERK), and IRE1α. These proteins activate the UPR, which is believed to be essential for viral replication.
ER stress induces kinase and ribonuclease (RNase) activities in IRE1α that lead to the excision of a 26 nucleotide intron in XBP1 mRNA. In addition, spliced XBP1 (XBP1s) upregulates genes that expand the ER lumen and its protein folding machinery.
Study findings
The authors observed that the four beta CoVs of HCoV-OC43, MERS-CoV, SARS-CoV-2, and MHV activate the IRE1α in host cells following infection. It was noted that except for SARS-CoV-2, the other CoVs induced splicing of XBP1 mRNA.
Further, all CoVs, aside from SARS-CoV-2, showed the induction of UPR-related genes. The researchers also demonstrated that both MERS-CoV and SARS-CoV-2 replicate in iAT2 cells and trigger activation of IRE1α.
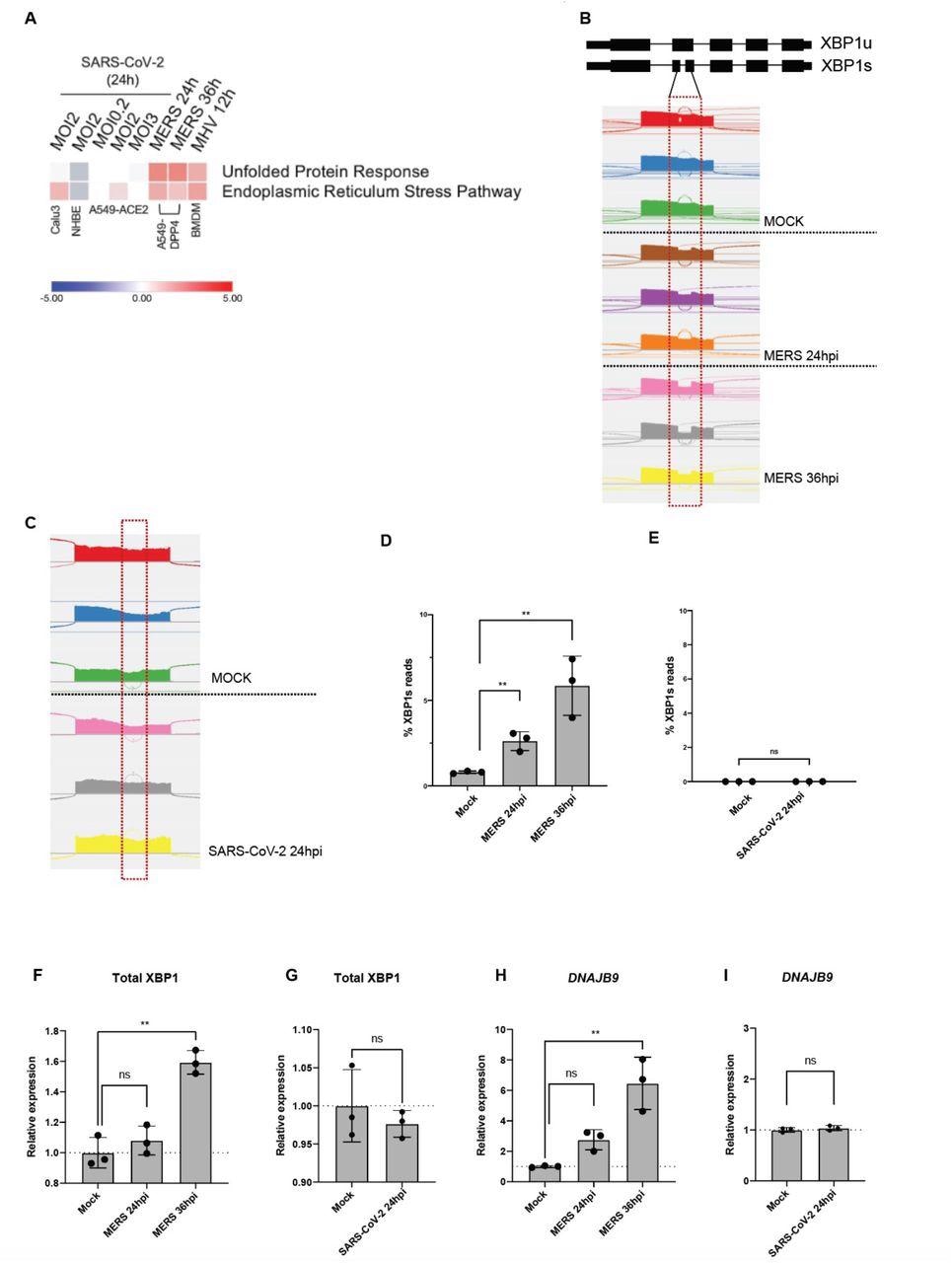
Unlike other coronaviruses, SARS-CoV-2 infection does not lead to robust UPR activation. (A) Heatmap of predicted pathway status based on Ingenuity Pathway Analysis (IPA) of activation z-scores for each pathway from RNA-sequencing data from indicated cells infected with SARS-CoV-2, MERS-CoV or MHV under specified conditions. Red: pathway predicted to be activated. Blue: pathway predicted to be inhibited. White: pathway predicted to be unchanged. Gray: no prediction due to lack of significance. (B&C) Quantification of XBP1 splicing by analyzing RNA-Seq data from A549-DPP4 and A549-ACE2 cells mock-infected or infected with MERS-CoV or SARS-CoV-2, respectively, under indicated conditions. Reads representing spliced or unspliced XBP1 mRNA were identified based on the presence or absence of the 26 nucleotide intron and quantified. (D-I) Percentage of XBP1 spliced reads, or relative expression of total XBP1 and DNAJB9 mRNA from the RNA-seq samples were then plotted. Values are means ± SD (error bars). Statistical significance was determined by Unpaired t-tests (* = P < 0.05; ** = P < 0.01; ns = not significant).
However, SARS-CoV-2 infection failed to induce splicing of XBP1 in these cells. It was found that SARS-CoV-2 causes active inhibition of the RNase activity of IRE1α.
The researchers sought to determine whether IRE1α was required for the replication and propagation of the four CoVs. To do this, the authors knocked out IRE1α by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing in A549 cell lines, which expressed receptors for each CoV.
All four CoVs were able to replicate in the cells lacking IRE1α, and the knock-out of IRE1α had no significant effects on their replication. This finding demonstrates that IRE1α is not essential for CoV replication.
A similar knock-out study was conducted for XBP1 in A459 cell lines. Likewise, no significant effect was noted. These findings provide evidence that none of the CoVs involved in the present study require the activation of IRE1α or XBP1 for replication.
Conclusions
HCoV-OC43, MERS-CoV, and MHV were found to activate the canonical IRE1α/XBP1 pathway in both A549 and Calu-3 cell lines. It was also reported that MERS-CoV could activate the IRE1α/XBP1 pathway in iAT2 cells.
The study noted that SARS-CoV-2 could partially activate the IRE1α/XBP1 pathway, as evidenced by autophosphorylation of IRE1α and the lack of splicing of XBP1. This suggests that the RNase activity of IRE1α was inactive.
In conclusion, the researchers observed that SARS-CoV-2 induces kinase activity of IRE1α but actively inhibits the RNase activity of the autophosphorylated IRE1α through an unknown mechanism, which the authors proposed as a probable strategy to evade detection by the host immune cells.
These findings present evidence that SARS-CoV-2 produces distinct changes in the activation of IRE1α following infection, which differs from that of other beta coronaviruses. Furthermore, the lack of IRE1α in host cells does not necessarily inhibit the replication of any of the beta CoVs tested.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Nguyen, L. C., Renner, D. M., Silva, D., et al. (2021). SARS-CoV-2 diverges from other betacoronaviruses in only partially activating the IRE1a/XBP1 ER stress pathway in human lung-derived cells. bioRxiv. doi:10.1101/2021.12.30.474519. https://www.biorxiv.org/content/10.1101/2021.12.30.474519v1.full.
- Peer reviewed and published scientific report.
Nguyen, Long C., David M. Renner, Diane Silva, Dongbo Yang, Nicholas A. Parenti, Kaeri M. Medina, Vlad Nicolaescu, et al. 2022. “SARS-CoV-2 Diverges from Other Betacoronaviruses in Only Partially Activating the IRE1α/XBP1 Endoplasmic Reticulum Stress Pathway in Human Lung-Derived Cells.” Edited by Diane E. Griffin. MBio 13 (5). https://doi.org/10.1128/mbio.02415-22. https://journals.asm.org/doi/10.1128/mbio.02415-22.