The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) consists of four main structural proteins including the spike, nucleocapsid, membrane, and envelope proteins. To date, most research on SARS-CoV-2 has focused on the effects and changes present in these proteins, as they are essential for the survival and pathogenesis of SARS-CoV-2 infection and are highly visible, thus making them excellent drug/vaccine targets.
However, several other SARS-CoV-2 proteins play a role in the survival of this virus during infection. To this end, researchers in a recent bioRxiv* study discuss the effects of the SARS-CoV-2 non-structural protein 1 (Nsp1), which binds to the ribosomal subunit thus inhibiting translation.
Study: Parsing the role of NSP1 in SARS-CoV-2 infection. Image Credit: Naeblys / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study findings
Previous studies have shown that SARS-CoV-2 Nsp1can inhibit translation and promote ribonucleic acid (RNA) degradation, whereas residues R124 and K125 are required for the inhibition of translation. To further assess whether Nsp1 is also necessary for messenger RNA (mRNA) degradation, two Nsp1 mutants were generated.
For the generation of Nsp1-deltaRB, amino acids 155-165 were removed, whereas the development of Nsp1-CD required both R124 and K125 to be mutated to alanine. Nsp1-deltaRB failed to be associated with ribosomes, while Nsp1-CD could associate but much less effectively than wild-type (WT).
Interestingly, the level of Nsp1 was lower for WT than the two mutants, thus indicating that it inhibits itself. When the expression of green fluorescence protein (GFP) reporters either fused to host betaglobin 5’UTR or the 5’ leader sequence of SARS-CoV-2, the WT and Nsp1-CD could both inhibit GFP expression in the host, but not in the leader sequence.
To further examine the effect of Nsp1 on translation mediated by an internal ribosome entry site (IRES), a bicistronic mRNA that allows expression of two luciferases to be mediated by either cap-dependent translation or downstream encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) was used. As expected, the WT Nsp1 was the most effective at suppressing both luciferases, with Nsp1-CD showing a lesser effect and Nsp1-deltaRB showing no effect.
To characterize the cellular transcripts sensitive to this degradation, the CoV2-5’ leader sequences upstream of the Nsp1 constructs were fused to prevent self-inhibition, which showed that WT with the CoV2-leader was refractory to its self-inhibitory effects and that the mutated construct increased expression.
SLAM-seq, which is an approach that allows for the measurement of endogenous mRNA, was then used to measure the induction of mRNA decay by Nsp1. This assay revealed U- to C- mutation rates at 1.5% that subsequently rose to 5.5%.
Cells expressing WT-Nsp1 also showed a more rapid turnover of RNA as both time and the portion of labeled RNA increased. Cells expressing Nsp1-deltaRB or Nsp1-CD showed no reduction in cellular mRNA half-lives.
A SARS-CoV-2 mutant (CoV2mut) was generated with the same amino acids (155-165 of nsp1) deleted. Vero and Calu3 cells were then infected with both WT SARS-CoV-2 and the mutant.
After confirming the expression of Nsp1 was not altered, the propagation of the virus was examined. Both cell types could support both viruses, with similar viral titers in Vero cells but much lower titers for the mutant in Calu3 cells.
This finding suggests that Nsp1 helps SARS-CoV-2 survive in the immunologically active Calu3 cells and indicates that Nsp1 interference could affect the interferon (IFN) response. To confirm this, a JAK-STAT inhibitor was used to prevent IFN signaling, which successfully rescued CoV2-mut propagation in Calu3 cells.
To further investigate the role of Nsp1 in SARS-CoV-2 pathogenesis, a Syrian hamster model was used. Two groups of eight hamsters were infected with either WT-SARS-CoV-2 or CoV2-mut and were weighed daily, with viral lung titers quantified after day seven.
CoV2-mut-infected mice exhibited less weight loss that became statistically significant at day six. In addition to reduced weight loss, CoV2-infected mice also exhibited significantly lower levels of viral lung titers that were below the limit of detection at seven days post-infection (dpi).
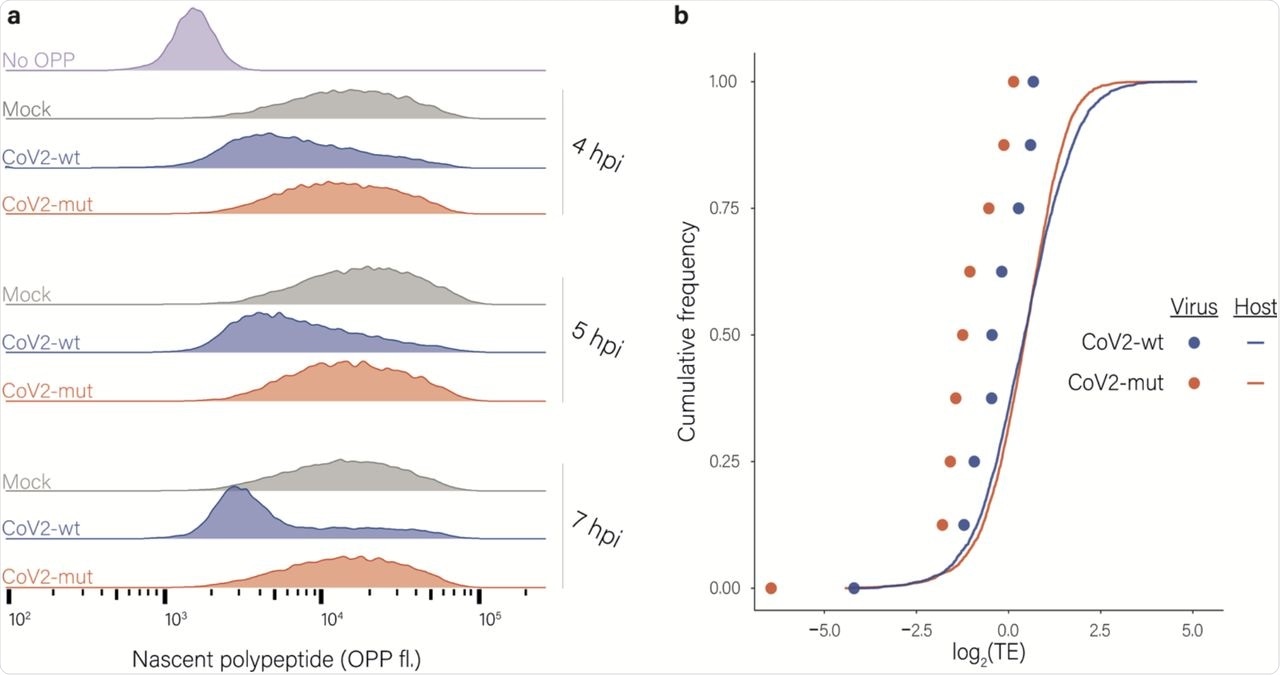
Effects of nsp1 on translation and accumulation of viral and host mRNAs. a, Protein synthesis measurement by flow cytometry of Calu3 cells infected with CoV2-wt or with CoV2-mut (MOI = 3) for 4, 5 and 7 hpi or an uninfected control following O-Propargyl Puromycin (OPP) incorporation and fluorescent labeling using Click chemistry b, Cumulative frequency of human (line) and viral (dots) genes according to their relative translation efficiency (TE) in cells infected with CoV2-wt (blue) or with CoV2-mut (red) at 4 hpi. TE was calculated from ribosome profiling and mRNA sequencing, and is defined as the ratio of ribosome footprints to mRNA for a given gene. Each dot represents one of nine major viral mRNA species.
Conclusions
The authors have successfully shown that nsp1 inhibits both capped and IRES-driven translation. Furthermore, the researchers here identified the residues that are essential to these processes and demonstrated that Nsp1 is vulnerable to inhibiting itself, which can be prevented to increase expression.
Further analysis demonstrated that Nsp1 RNA degradation is dependent on ribosome binding. Investigations in Vero and Calu3 cells proved that Nsp1 is important for viral survival, that the mRNA degradation effects remain active during infection, that the loss of this protein in vivo alters the pathogenicity of SARS-CoV-2 infection.
Taken together, the findings from this study help identify Nsp1 is a key protein for the survival and effectiveness of SARS-CoV-2. In the future, this information could assist researchers in developing novel therapeutics to reduce the severity or length of the disease in coronavirus disease 2019 (COVID-19) patients.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fisher, T., Gluck, A., Narayanan, K., et al. (2022). Parsing the role of NSP1 in SARS-CoV-2 infection. bioRxiv. doi:10.1101/2022.03.14.484208. https://www.biorxiv.org/content/10.1101/2022.03.14.484208v2.full.
- Peer reviewed and published scientific report.
Fisher, Tal, Avi Gluck, Krishna Narayanan, Makoto Kuroda, Aharon Nachshon, Jason C. Hsu, Peter J. Halfmann, et al. 2022. “Parsing the Role of NSP1 in SARS-CoV-2 Infection.” Cell Reports 39 (11). https://doi.org/10.1016/j.celrep.2022.110954. https://www.cell.com/cell-reports/fulltext/S2211-1247(22)00736-7.