Several SARS-CoV-2 variants of concern have emerged since the beginning of the coronavirus diseases 2019 (COVID-19) pandemic. These variants differ from each other in characteristics, including transmissibility and infectivity. Various studies have reported that mutations in the viral spike protein are responsible for these alterations.
 Study: Shared N417-dependent epitope on the SARS-CoV-2 Omicron, Beta and Delta-plus variants. Image Credit: NIAID
Study: Shared N417-dependent epitope on the SARS-CoV-2 Omicron, Beta and Delta-plus variants. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the present study, researchers compared the cross-reactivity exhibited by plasma responses during different SARS-CoV-2 waves dominated by SARS-CoV-2 D614G, Beta, and Delta variants.
The team obtained plasma samples from patients infected in the DG14G-dominant period prior to September 2020, the Beta-dominant wave between December 2020 and January 2021, and the Delta-dominant wave in July 2021. The samples were collected from HIV-uninfected individuals aged more than 18 years.
The researchers first compared the antibody responses observed in unvaccinated individuals infected by either the D614, Beta, or the Delta variants between May 2020 and July 2021. The team also explored the neutralization ability of plasma elicited by the three variants and SARS-CoV-1. Furthermore, the geometric mean titers (GMTs) were compared for the corresponding plasma responses elicited by the SARS-CoC-2 variants and SARS-CoV-1 to evaluate the extent and patterns of cross-reactivity against each variant.
The team also assessed whether the antibodies to the N417K mutation in the Beta receptor-binding domain (RBD) were responsible for the plasma responses against the Beta variant. This was achieved by examining Beta-infected plasma samples and the Delta+ variant. Additionally, the team assessed the importance of N417 as a target of monoclonal antibody (mAb) responses elicited by Beta by isolating mAbs from a Beta-infected person who had displayed potent antibody responses towards Beta and cross-reactivity towards other variants.
The team subsequently mapped the S protein target of the mAb by conducting binding experiments with SARS-CoV-2-specific enzyme-linked immunosorbent assay (ELISA). The ability of the mAbs to facilitate antibody-dependent cellular phagocytosis (ADCP) was also analyzed. Moreover, the ability of the mAbs to crosslink SARS-CoV-2 S proteins was evaluated.
Results
The study results showed that plasma elicited by the SARS-CoV-2 D614G variant generated potent autologous neutralization of the D614G spike (S) protein. However, D614G-elicited plasma also displayed a 12- to 15-fold reduction in neutralization towards the SARS-CoV-2 Beta and Delta variants as well as SARS-CoV-1. In contrast, the team also observed that the plasma from Beta-infected individuals showed a 2.9-fold reduced neutralization of the D614G variant, suggesting significant cross-reactivity. However, the Beta-elicited plasma showed lower cross-reactivity, which was reduced by 11.3-fold against Delta, 9.4-fold against Omicron, and 9.3-fold against SARS-CoV-1.
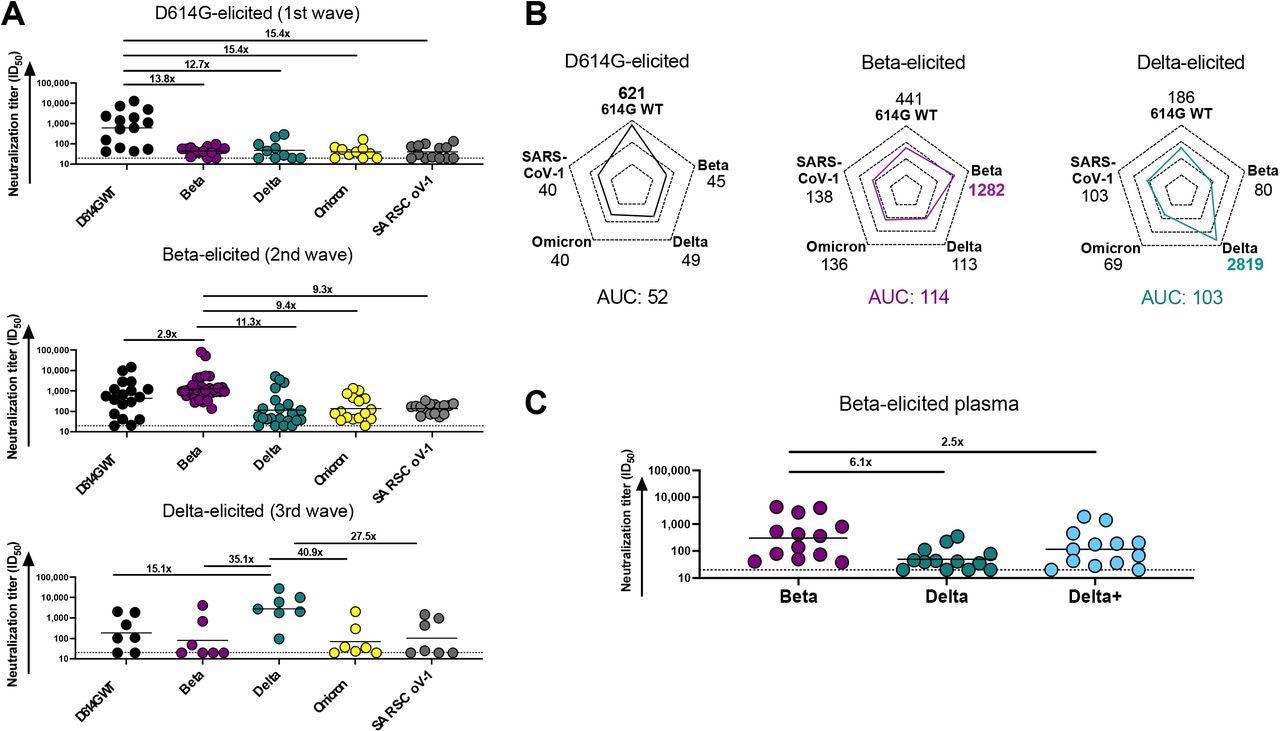
Comparison of plasma cross-reactivity elicited by three distinct SARS-CoV-2 variants. (A) Plasma from D614G, Beta and Delta infections during three distinct SARS-CoV-2 waves were tested for neutralization breadth against a range of VOCs using a pseudovirus-based neutralization assay. Fold changes in neutralization are shown above each variant. Plasma neutralization titer is measured as an ID50. Black horizontal bars represent geometric means. The threshold of detection for the neutralization assay is ID50>20. (B) Spider plots were derived from GMTs for plasma triggered by D614G, Beta or Delta against multiple VOCs. The GMTs for each plasma set were normalized against titers to the autologous virus and breadth was expressed as area under the curve. (C) Plasma from Beta-infected individuals was tested against the Delta and Delta+ variants. Fold changes in neutralization are shown above each variant. Plasma neutralization titer is measured as an ID50. Black horizontal bars represent geometric means. The threshold of detection for the neutralization assay is ID50>20.
Notably, while the plasma in Delta-infected individuals had remarkably high autologous titers against the matched S protein, the team observed a 15- to 41-fold reduction in neutralization potency against the Delta, Beta, and Omicron variants and SARS-CoV-1. Also, a high level of cross-reactivity was found against the D614G variant in Delta-elicited samples.
The team observed that each variant generated the highest plasma responses against their autologous S proteins. However, the degree of the autologous neutralization was different across different variants as Delta triggered the highest response, followed by Beta and D614G. Moreover, the plasma responses elicited by the Beta and the Delta variants were almost two times higher than those by the D614G variant. This indicated that the neutralization responses elicited by Beta and Delta were more cross-reactive as compared to those of D614G.
While Beta plasma had a 6.1-fold reduction in antibody responses against the Delta variant, a 2.5-fold decline in potency was observed against the Delta+ variant in the Beta samples. This highlights the significance of the N417 residue as a target of antibody responses elicited by the Beta variant. Furthermore, mAb obtained from Beta-infected individuals neutralized Beta and Delta+ pseudoviruses as well as the Omicron variant. However, these mAbs were not potent against the D614G, Delta, or the SARS-CoV-1 pseudoviruses.
The ELISA reported no detectable binding for either the D614G and Beta N-terminal domain (NTD) proteins; however, robust binding was found towards the Beta RBD. Notably, the mAb bound to the mutant K417N did not bind to the D614G subdomains. Altogether, this data indicated that mAb was substantially dependent on the N417 residue found in the Beta RBD.
Overall, the study findings showed that N417-dependent mAbs could serve as an excellent target to stimulate cross-reactive responses against SARS-CoV-2 variants. The researchers believe that the discovery of mAbs that target conserved viral sites will help develop therapeutic mAb combinations to treat SARS-CoV-2 infections, irrespective of the infecting variant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Shared N417-dependent epitope on the SARS-CoV-2 Omicron, Beta and Delta-plus variants, Thandeka Moyo-Gwete, Mashudu Madzivhandila, Nonhlanhla N Mkhize, Prudence Kgagudi, Frances Ayres, Bronwen E Lambson, Nelia P Manamela, Simone I Richardson, Zanele Makhado, Mieke van der Mescht, Zelda de Beer, Talita Roma de Villiers, Wendy A Burgers, Ntobeko AB Ntusi, Theresa Rossouw, Veronica Ueckermann, Michael T Boswell, Penny L Moore, medRxiv 2022.04.24.22273395, DOI: https://doi.org/10.1101/2022.04.24.22273395, https://www.medrxiv.org/content/10.1101/2022.04.24.22273395v1
- Peer reviewed and published scientific report.
Moyo-Gwete, Thandeka, Mashudu Madzivhandila, Nonhlanhla N. Mkhize, Prudence Kgagudi, Frances Ayres, Bronwen E. Lambson, Nelia P. Manamela, et al. 2022. “Shared N417-Dependent Epitope on the SARS-CoV-2 Omicron, Beta, and Delta plus Variants.” Edited by Tom Gallagher. Journal of Virology 96 (15). https://doi.org/10.1128/jvi.00558-22. https://journals.asm.org/doi/10.1128/jvi.00558-22.