Astroviruses are single-stranded, non-enveloped, positive-sense ribonucleic acid (RNA) viruses, possessing a genome of nearly 6,000 to 7,700 nucleotides. They are linked with infections of the nervous and gastrointestinal systems and exhibit further tissue tropisms. CoVs are single-stranded, enveloped, positive-sense RNA viruses and harbor a genome of about 30,000 nucleotides. In addition, the member of the CoV family, i.e., SARS-CoV-2, is responsible for the ongoing CoV disease 2019 (COVID-19) pandemic.
Prior studies have indicated that viruses belonging to distinct viral families, such as CoVs and astroviruses, harbor an RNA component named the s2m. It was discovered near the 3′ end of classic human astroviruses and avian bronchitis virus, a CoV, in 1997 by Monceyron et al. Existing reports indicated that the secondary form of the s2m is significant for virus multiplication. However, the functional importance of s2m in the viral lifecycle remains uncertain.
About the study
In the present study, the researchers analyzed the functional relevance of the s2m for the lifecycle of three viruses: SARS-CoV-2, astrovirus VA1 (VA1), and classic human astrovirus 1 (HAstV1). For this, the team mutagenized the s2m component employing reverse genetics. They also studied a clinical isolate of SARS-CoV-2 with a natural deletion in the s2m domain. Thus, the scientists generated substitutions or deletions of the s2m in SARS-CoV-2, VA1, and HAstV1.
The importance of s2m for viral propagation and guanine-cytosine (G-C) binding pairs for the functioning of s2m in VA1 was assessed. Likewise, the relevance of s2m in another astrovirus, HAstV1, was determined to corroborate the initial findings. Furthermore, the authors evaluated the role of s2m in SARS-CoV-2 in vitro and in vivo. They analyzed the growth of a clinical isolate of SARS-CoV-2 possessing a deletion in the s2m region.
Results
The study results showed that the s2m was essential for VA1 and HAstV1. The s2m mutation or deletion in SARS-CoV-2, on the other hand, did not affect the virus's ability to grow in vivo/in vitro. These findings revealed that the s2m's function and significance were virus-context reliant.
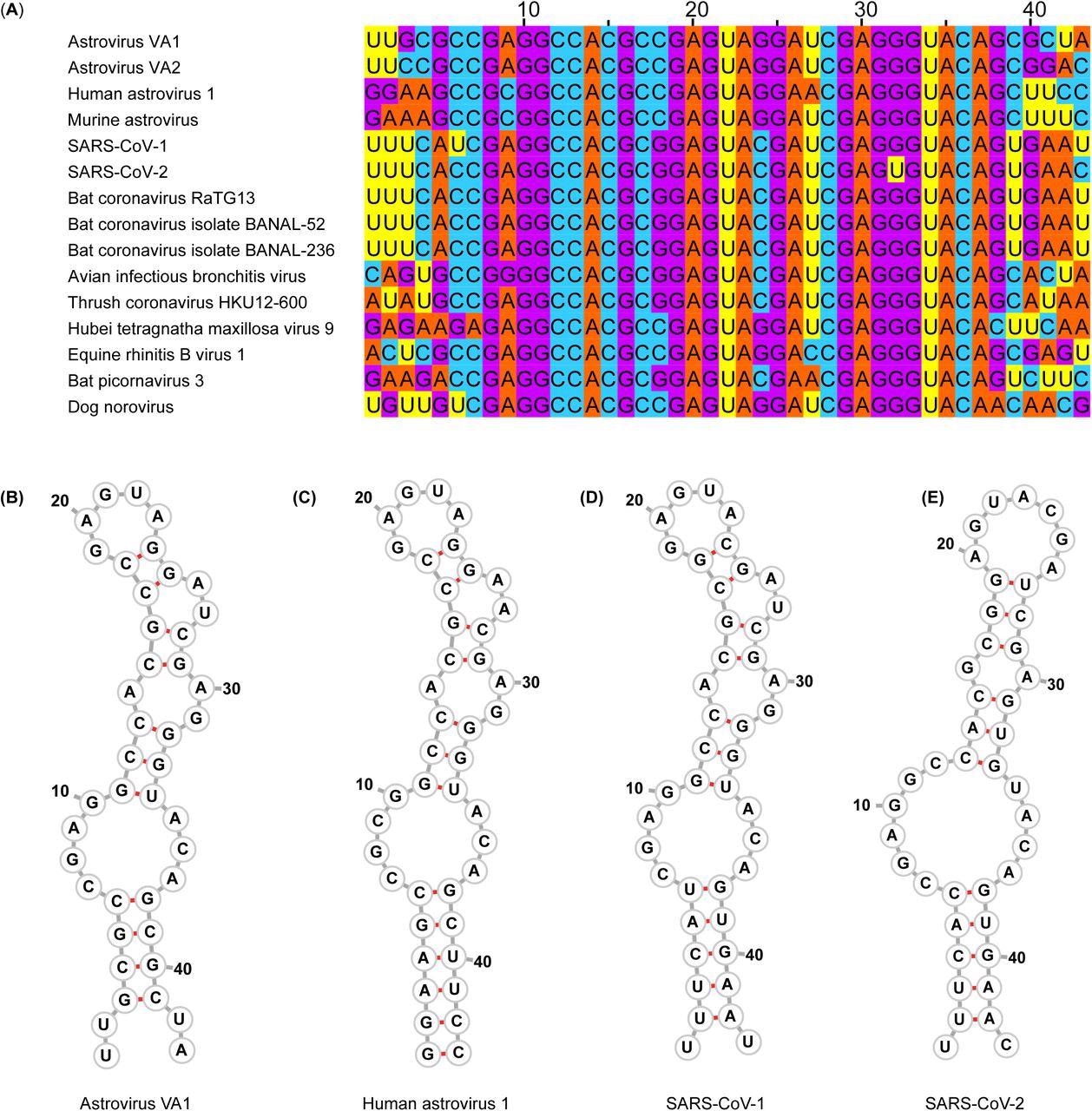
s2m nucleotide sequences and predicted secondary structures. (A) Multiple sequence alignment of representative viruses that encode an s2m. (B-E) Predicted secondary structure of the s2m by RNAfold for (B) Astrovirus VA1, (C) Human astrovirus 1, (D) SARS-CoV-1, (E) SARS-CoV-2
The findings for VA1 showed that G-C binding pairs were crucial in the s2m element, confirming the hypothesized tertiary and secondary structure. The need to preserve the s2m's structure presumably limits the nucleotide range at each position. While position 24 was preserved with a complementary base that retains a G-C binding pair, position 18 could be G or C. VA1 tolerated all orientations of this G-C base pair with no diminution in viral progeny.
Until the anticipated structure of s2m was preserved, VA1 tolerated interruption of the stop codon and the incorporation of 16 amino acids to the C-terminus of open reading frame 2 (ORF2). Nucleotide locations that were not anticipated to generate G-C base pairs could be altered in the s2m, although this reduces viral progeny generation.
Additionally, the authors corroborated the importance of the s2m in another astrovirus, HAstV1, and discovered that it was crucial but not vital. These data show that the s2m's significance was determined by the virus.
Further, the scientists noted that the VA1 genetic variant with uracil (U) at position 32 with s2m could not be rescued. This result confirmed the importance of G-C base pairs in sustaining the s2m structure. It was also in line with the finding that in SARS-CoV-2, the s2m element was not required. Moreover, SARS-CoV-2 harbored U at position 32, whereas all other s2m-containing viruses encoded G.
The present results illustrated that the SARS-CoV-2 s2m was non-essential for the viral lifecycle in vivo/in vitro. The authors speculated that SARS-CoV-2 might have evolved a method to replace the s2m's function. Alternatively, the SARS-CoV-2 s2m might possess a unique purpose but still be unnecessary. Since the U form of SARS-CoV-2 was conserved throughout clinical isolates, it may give an anonymous fitness benefit.
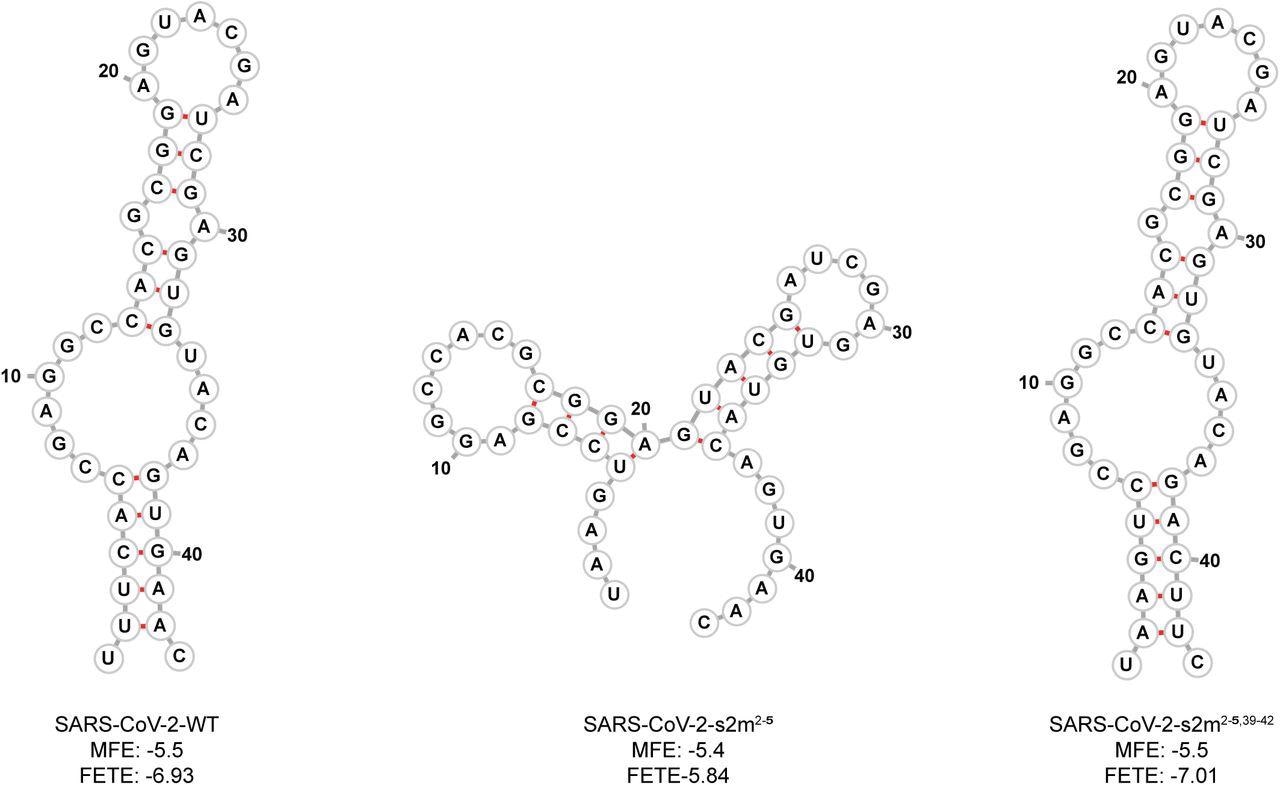
Predicted SARS-CoV-2 WT and mutant s2m secondary structures. Using RNAfold, the predicted minimum free energy secondary structure of the WT and mutant s2m RNA sequences substituted into the SARS-CoV-2 genome are shown. The calculated minimum free energy (MFE) and free energy of the thermodynamic ensemble (FETE) of each sequence are also shown.
Conclusions
The authors stated that despite the initial presentation of the s2m component in 1997, no functional genetic research on the element's role in the viral lifecycle existed.
The current study showed that after partial deletion of the s2m or replacements of G-C base pairs, the recombinant virus for VA1 could not be recovered. Compensatory substitutions restoring the G-C base pair allowed the recovery of VA1. A partial deletion of the s2m in HAstV1 led to lower viral titers relative to a wild-type virus and less activity in a replicon system. On the contrary, the capacity to rescue the virus, and growth in vitro and growth in Syrian hamsters, were unaffected by mutation or deletion of the SARS-CoV-2 s2m.
Overall, the present study illustrated the relevance of the s2m was virus-dependent. The investigators mentioned that future studies should focus on analyzing the mechanistic reason why a highly conserved RNA component might lead to diverse functional activities in the viral lifecycle.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
The highly conserved stem-loop II motif is important for the lifecycle of astroviruses but dispensable for SARS-CoV-2; Andrew B Janowski, Hongbing Jiang, Chika Fujii, Macee C Owen, Traci L Bricker, Tamarand L Darling, Houda H Harastani, Kuljeet Seehra, Stephen Tahan, Ana Jung, Binita Febles, Joshua A Blatter, Scott A Handley, Bijal A Parikh, Valeria Lulla, Adrianus CM Boon, David Wang. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.04.30.486882, https://www.biorxiv.org/content/10.1101/2022.04.30.486882v1