A groundbreaking study reveals how a single gene variant shapes the immune landscape of the brain and blood, unlocking new opportunities for early detection and intervention across Alzheimer’s, Parkinson’s, and other neurodegenerative disorders.
 Study: APOE ε4 carriers share immune-related proteomic changes across neurodegenerative diseases. Image Credit: Andrii Vodolazhskyi / Shutterstock
Study: APOE ε4 carriers share immune-related proteomic changes across neurodegenerative diseases. Image Credit: Andrii Vodolazhskyi / Shutterstock
In a recent study published in the journal Nature Medicine, researchers identified a conserved immune signature associated with the apolipoprotein E ε4 variant (APOE ε4) across cerebrospinal fluid (CSF), brain, and plasma, regardless of the presence of neurodegenerative disease.
The APOE ε4 gene is the most significant genetic risk factor for late-onset Alzheimer’s disease (AD). However, mounting evidence suggests APOE ε4 carriage may play a role in other neurodegenerative diseases. APOE ε4 has been linked to a lower age of onset and a higher risk of Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD). APOE ε4 has also been linked to poor cognition and faster cognitive decline in PD, elevating PD dementia (PDD) risk.
Notwithstanding APOE ε4’s deleterious impact, little is known about its biological mechanisms and whether and how it changes across neurodegenerative diseases. The paper frames this within the evolutionary concept of "antagonistic pleiotropy," suggesting that the gene’s pro-inflammatory effects may be beneficial against infectious diseases in youth but become harmful with age. Previously, the authors reported that APOE ε4 carriers shared a pro-inflammatory proteomic signature in the CSF regardless of cognitive status in mild cognitive impairment and AD. Whether this applies to other neurodegenerative diseases remained unknown.
The study and findings
The present study explored systemic proteomic changes in APOE ε4 carriers with neurodegenerative diseases. First, SomaScan proteomic data from the Global Neurodegeneration Proteomics Consortium (GNPC) dataset, which reflects real-world clinical heterogeneity, were utilized for the CSF proteome profiling of individuals with AD, PD, or control (non-impaired) status. Using mutual information, 229 CSF proteins associated with APOE ε4 were identified in non-impaired controls.
Classification and regression trees (CART) models showed that these 229 proteins could differentiate APOE ε4 carriers from non-carriers across PD and AD. A functional enrichment analysis of CSF APOE ε4 proteins indicated significant enrichment for apoptosis, viral processes, protein folding and phosphorylation, RNA/DNA processes, cellular processes, and rhythmicity.
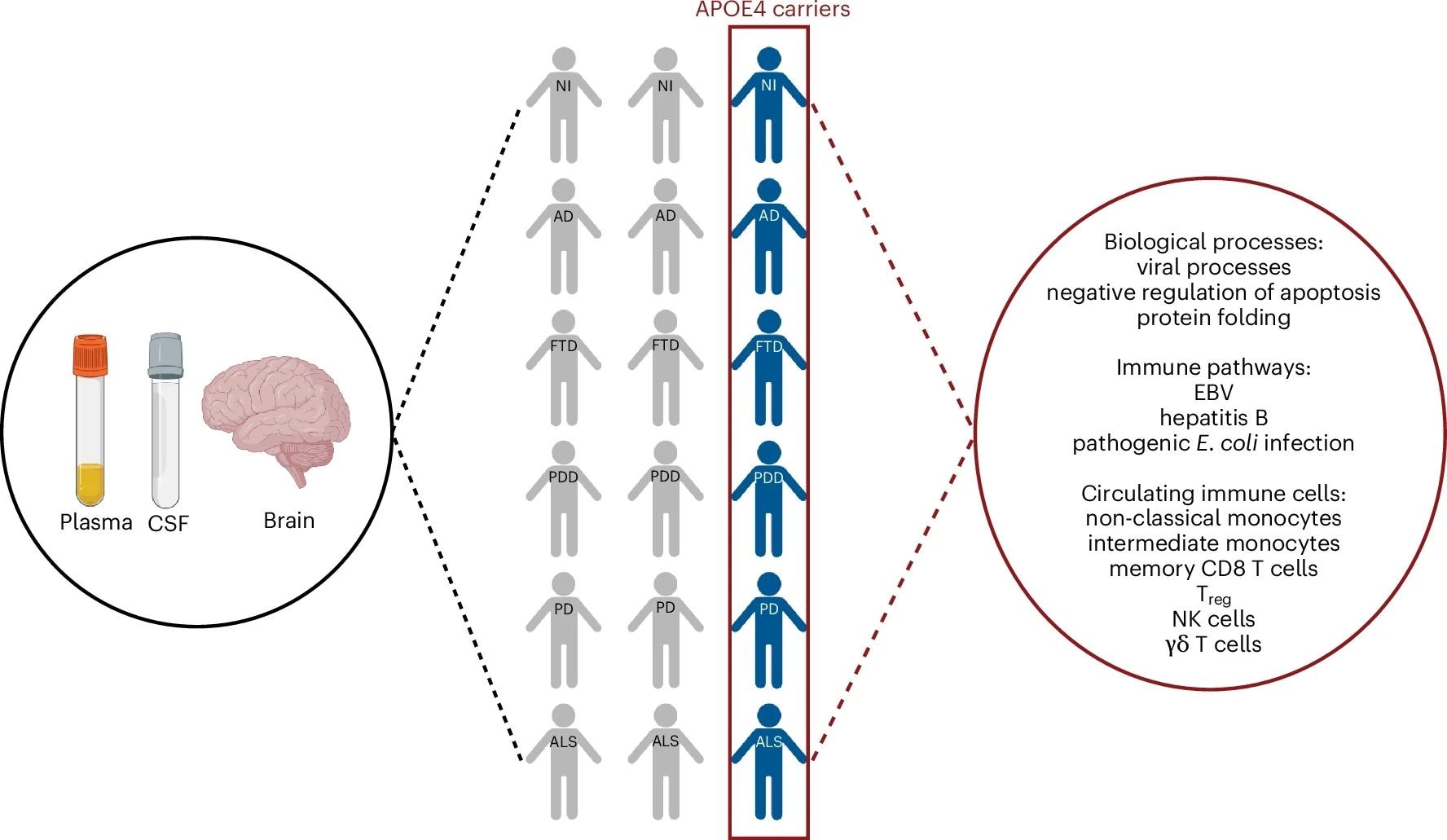 APOE ε4 carriers across different neurodegenerative diseases share a common systemic proteomic change reflective of pro-inflammatory immune dysregulation. The figure is created with BioRender.com.
APOE ε4 carriers across different neurodegenerative diseases share a common systemic proteomic change reflective of pro-inflammatory immune dysregulation. The figure is created with BioRender.com.
Further analysis for immune-specific processes revealed APOE ε4 enrichment in various infection-related pathways, including herpes, influenza A, hepatitis, measles, and Epstein-Barr virus (EBV). Significant enrichment was also observed for B-cell, T-cell, and inflammatory signaling cascades. Next, immune cell subtype enrichment analysis revealed the most APOE ε4 enrichment in intermediate and non-classical monocytes among innate immune cells.
Among adaptive immune cells, memory cluster of differentiation 8 (CD8) T cells, regulatory T (Treg) cells, and memory CD4 T cells were the most enriched. Besides, γδ T cells and natural killer (NK) cells showed APOE ε4 enrichment. In the liver, a cell-type-specific enrichment analysis revealed the most APOE ε4 enrichment in Kupffer cells and hepatocytes.
Next, the researchers examined whether APOE ε4 CSF proteome changes were reflected in the plasma and used the GNPC dataset for plasma proteome profiling of AD, PDD, FTD, PD, ALS, and non-impaired controls. Fifty-eight plasma proteins associated with the APOE genotype were identified in non-impaired controls. CART modeling revealed that these 58 proteins could strongly differentiate between APOE ε4 carriers and non-carriers across neurodegenerative diseases, and this signature was found to be consistent across different sexes and racial groups.
APOE ε4 plasma processes showed significant enrichment in biological processes, including protein processes, cellular processes, viral processes, DNA/RNA processes, and apoptosis. Viral processes were the most significantly enriched in both plasma and CSF. This was supported by similar enrichments in infection and immune pathways, including hepatitis and EBV. Intermediate and non-classical monocytes and basophils were enriched for APOE ε4 proteins among innate immune cells.
NK cells, γδ T cells, memory CD8 T cells, and naïve CD8 T cells were enriched for APOE ε4 proteins among adaptive immune cells. In the liver, T cells and Kupffer cells were mainly enriched, while hepatocytes showed little enrichment. These data indicate that genotype-specific proteomic changes detected in the CSF were also reflected in the plasma in APOE ε4 carriers and non-carriers.
Further, the team explored whether peripheral proteomic changes were mirrored in the brains of APOE ε4 carriers and non-carriers. To this end, they utilized the proteomic data of the dorsolateral prefrontal cortex (dlPFC) from individuals with AD, FTD, PD, PDD, ALS, and non-impaired individuals, as part of the Accelerating Medicines Partnership for AD (AMP-AD) UPenn Proteomics Study. Using mutual information, 248 APOE ε4 proteins were identified in the dlPFC.
Functional enrichment analyses showed that the three main biological processes (viral processes, apoptosis, and protein folding) identified in the plasma and CSF were also significantly enriched in the dlPFC of APOE ε4 carriers across neurodegenerative diseases. Four of the most significantly enriched immune pathways in the plasma and CSF were also identified in APOE ε4 carriers disease-independently: hepatitis B, EBV, Escherichia coli infection, and viral carcinogenesis. Crucially, the study found that this brain signature was independent of the hallmark neurodegenerative pathologies, including amyloid-β, tau, TDP-43, and α-synuclein, reinforcing that the APOE ε4 effect is a fundamental vulnerability rather than a direct result of disease-specific protein aggregation.
An Unexpected Finding Challenges a Key Biomarker
In a notable and "unexpected finding," the study revealed that plasma neurofilament light (NEFL) levels, a widely used biomarker of neurodegeneration, were consistently lower in APOE ε4 carriers. The authors note this contradicts some prior research and "raises important questions about the reliability of NEFL as a stand-alone biomarker," suggesting its levels may be influenced by APOE ε4-related metabolic factors or clearance across the blood-brain barrier.
Finally, a correlation network analysis was performed to investigate the relationship between APOE ε4 proteins and clinical, demographic, and lifestyle factors in PDD, PD, AD, and non-impaired APOE ε4 carriers. They identified disease-specific relationships with APOE in neurodegenerative diseases. APOE was associated with race and sex in AD, diabetes in PD, age, hypertension, and body mass index in non-impaired controls, and chronic obstructive pulmonary disease and resting heart rate in PDD. The authors caution, however, that because the study is cross-sectional, they "cannot infer causality," as these clinical factors could be a consequence of the neurodegenerative disease rather than a cause.
Conclusions
The findings showed that APOE ε4 carriers share a unique proteomic signature across the plasma, CSF, and brain, regardless of neurodegenerative disease. This signature was associated with enrichment for circulating immune cells and pro-inflammatory immune dysregulation. The authors propose a specific mechanism for this, suggesting that hyperactive peripheral immune cells may interact with and disrupt the blood-brain barrier (BBB), thereby driving neuroinflammation. However, proteins within this signature were uniquely correlated with clinical, demographic, and lifestyle factors in a neurodegenerative disease-specific manner.
This suggests that APOE ε4 confers a systemic biological vulnerability that is essential but insufficient for neurodegeneration, underscoring the need to account for gene-environment interactions. The authors also acknowledge limitations, including the "absence of validated biomarkers" to confirm all clinical diagnoses and the "absence of direct measures of routine inflammatory markers such as C-reactive protein," which should be addressed in future studies.
Overall, the results reframe APOE ε4 as a pleiotropic immune modulator, rather than an AD-specific risk gene, providing a foundation for early intervention strategies and precision biomarker development across neurodegenerative diseases. The study concludes by calling for a conceptual shift in the field, moving from simply identifying genetic risk loci "toward functional characterization of established variants," and establishing a roadmap for future research into these complex interactions.