The S100 family is a set of dimeric, calcium-binding proteins which have been seen to be over expressed in several types of cancer. S100 proteins attach to the Cterminal region (293-393) of p53, the most significant tumor suppressor involved in cell cycle arrest, DNA repair and apoptosis. As S100 is dimeric and the C-terminus of p53 includes a tetramerization domain, the potential range of complexes integrating S100 and p53 is large.
p53 tetramer interacting with DNA. Source: PDBe 3kz8
Featuring a huge fraction of intrinsically disordered structure, the p53 variants do not elute as per globular protein standards in size-exclusion chromatography (SEC). However, multi-angle light scattering finds molecular mass based on first principles and does not rely on SEC column calibration standards. The combination of SEC and static multi-angle light scattering (SEC-MALS) offers a perfect system to investigate the complex formation of S100 proteins and p53; it ascertains the molecular weight of the complexes independently of retention time and protein standards, under native buffer conditions. SEC-MALS was used to study how S100 proteins attach to p53 in its varied oligomeric states, using dimeric (L344A) and monomeric (L344P) p53 mutants.
Materials and Methods
The experimental system included a SEC and HPLC column connected in series with an Optilab® differential refractive index (dRI) detector and a DAWN® MALS detector. Data collection and analysis were carried out in the software. MALS combined with dRI®ASTRA concentration data is sufficient to compute the weight-average molar mass of solution at each elution volume in the chromatogram, as plotted in the figures.
The S100 proteins and p53 mutants were initially characterized separately by SEC-MALS to establish their homogeneity and molecular masses. Later, they were incubated at different stoichiometric ratios—1:1, 1:2, 1:4, 2:1, and 4:1—and examined on the SEC-MALS system.
<h2>Results and Discussion
In Figure 1, molar mass results of the individual p53 proteins are superimposed with the light scattering (thin solid lines) and dRI chromatograms (dotted lines). The molar masses correspond to a tetramer for p53 wild-type, a dimer for the L344A variant, and a monomer of 11.2 kDa for the L344P variant. Slight curvature of the wild-type and L344A mutant molar mass plots corresponds with concentration within the peak, denoting a small degree of dynamic equilibrium at these concentrations. The monomeric L344P mutant is relatively homogeneous across its peak.
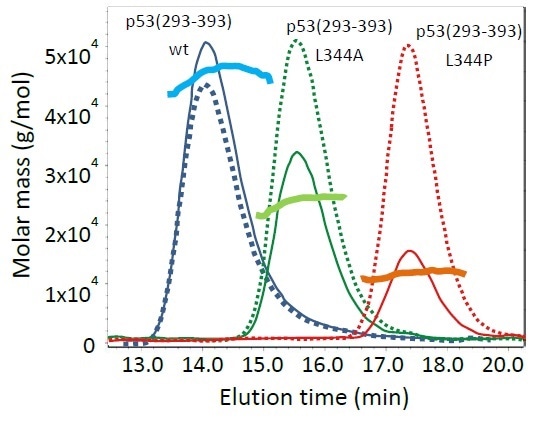
Figure 1. Analytical SEC-MALS of p53(293-393) variants. Thin, solid lines are light scattering chromatograms, dotted lines the corresponding dRI signals and thick solid lines are the calculated molar masses.
Figure 2 illustrates the light scattering chromatograms and calculated molar masses for the pre-incubated mixtures. Both monomeric p53 (293-393) L344P (11.2 kDa) and dimeric S100B (21.4 kDa) were eluted with a retention time of ~17.5 minutes. The complex molar mass of ~32 kDa at ~16 minutes do not depend on the relative ratio of the two proteins, signifying that one dimer of S100B binds only a single monomer of p53. Again, concentration-dependent curvature of the molar mass in the complex peak denotes rapid equilibration of monomers and complexes during the elution, a property not apparent in standard SEC.
Table 1. Summary of oligomers and protein-protein complexes found. Stoichiometry refers to the number of each monomer in the complex.
| |
S100B |
w.t. |
L344A |
L344P |
| Native oligomer: |
dimer |
tetramer |
dimer |
monomer |
| Stoichiometry |
Complex forms with S100B? |
| 1:1 |
|
- |
- |
- |
| 2:1 |
|
- |
√ |
√ |
| 2:2 |
|
- |
- |
- |
| 4:1 |
|
- |
- |
- |
| 8:4 |
|
√ |
- |
- |
In additional data which is not shown in this article, it was found that no complexes containing two monomers of wild-type p53 (~43 kDa) and a dimer of S100B, or of two dimers binding one monomer of p53 (~54 kDa), could be identified. However, S100B was able to form a complex with the monomer by disrupting the dimeric p53 (293- 393) L344A mutant. Interestingly, SEC-MALS showed that S100B was also able to form a stable complex with tetrameric w.t. p53 (293-393); the complex’s molar mass comprised of 4 dimers of S100B attached to a tetramer of p53.
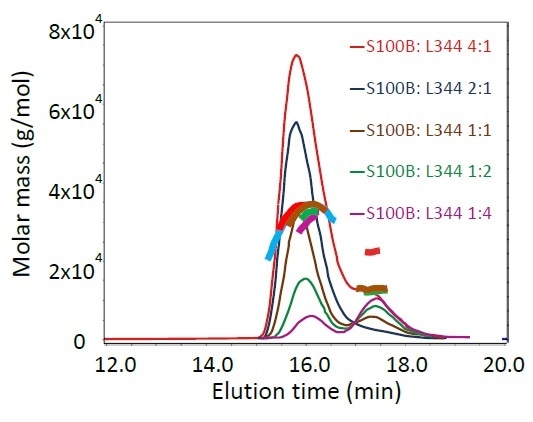
Figure 2. SEC-MALS results of S100B and the mutant p53(293- 393)L344P, pre-incubated in different stoichiometric ratios: light scattering chromatograms and calculated molar masses across each peak. Curvature of the molar masses at the 16- minute peak indicates dynamic equilibrium of the complex.
Conclusions
SEC-MALS helped to understand the way S100 proteins affect the oligomerization of p53. Based on these results and in vivo studies, a binding model was established where S100 proteins is capable of activating tetrameric p53 but inhibits p53 activity binding to the monomer by shifting the tetramerization equilibrium. Thus, SEC-MALS is an important tool in structural biology for analyzing the stoichiometry and formation of several types of protein-protein complexes.
About Wyatt
 With a long history of excellence in scientific instrumentation, Wyatt Technology is the recognized leader in innovative light scattering instruments, accessories, software and services for determining the properties of macromolecules and nanoparticles in solution. Wyatt provides cutting-edge solutions for in-line multi-angle static light scattering (SEC-MALS), field-flow fractionation (FFF-MALS), composition gradients (CG-MALS), high-throughput and traditional dynamic light scattering (DLS), electrophoretic mobility via phase-analysis light scattering (MP-PALS), differential refractometry and differential viscosity. With a staff composed of 20% Ph.D. scientists and many more dedicated and experienced support personnel, Wyatt's aim is to delight the customer with the best products, training, customer support and service available in the industry.
With a long history of excellence in scientific instrumentation, Wyatt Technology is the recognized leader in innovative light scattering instruments, accessories, software and services for determining the properties of macromolecules and nanoparticles in solution. Wyatt provides cutting-edge solutions for in-line multi-angle static light scattering (SEC-MALS), field-flow fractionation (FFF-MALS), composition gradients (CG-MALS), high-throughput and traditional dynamic light scattering (DLS), electrophoretic mobility via phase-analysis light scattering (MP-PALS), differential refractometry and differential viscosity. With a staff composed of 20% Ph.D. scientists and many more dedicated and experienced support personnel, Wyatt's aim is to delight the customer with the best products, training, customer support and service available in the industry.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.