This article is based on a poster originally authored by Dagmara Wanda Lewandowska, PhD, Marc Gasser, PhD, Vanina Haunreiter-Dengler, PhD and Eva Meszaros, PhD, which was presented at ELRIG Drug Discovery 2024 in affiliation with INTEGRA Biosciences and Microsynth AG.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
This study shows the comparable performance of INTEGRA Biosciences‘ MAGFLO NGS magnetic beads with gold standard magnetic beads when used for size selection in next-generation sequencing (NGS) library preparation, followed by Illumina MiSeq amplicon sequencing. Amplicons of 16S rRNA and internal transcribed spacer (ITS) were obtained from a microbial community standard containing genomic DNA from 8 bacterial species (3 Gram-negative and 5 Gram-positive) and 2 yeasts. The study demonstrates no significant difference between the performance of MAGFLO NGS and gold standard magnetic beads during size selection steps in library preparation or in consecutive sequencing results and no sequencing bias in alpha diversity analysis of microbial composition.
Introduction
Size selection with magnetic beads is the gold standard technique for NGS library preparation. It is used for either single-size selection in PCR clean-up or double-size selection of fragments within a desired size range for sequencing. Reproducible and efficient size selection ensures consistent sequencing results.

Figure 1. Amplicons for 16S and ITS2 were generated using the ZymoBIOMICS™ Microbial Community DNA Standard (D6305) with standard primers. Magnetic bead clean-up and size selection steps during the library preparation protocol were performed either with gold standard or MAGFLO NGS beads (indicated by color). Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Table 1. The theoretical composition of 16S (or 16S and 18S) rRNA gene abundance was used as a reference, and was calculated from the theoretical genomic DNA composition using the following formula: 16S/18S copy number = total genomic DNA (g) × unit conversion constant (bp/g) / genome size (bp) × 16S/18S copy number per genome. This should be used as a reference when performing 16S targeted sequencing3. Source: ELRIG (UK) Ltd.
| Species |
Theoretical composition (%) |
Genomic
DNA |
16S
only |
16S &
18s |
Genome
copy |
Cell
number |
| Pseudomonas aeruginosa |
12 |
4.2 |
3.6 |
6.1 |
6.1 |
| Escherichia coli |
12 |
10.1 |
8.9 |
8.5 |
8.5 |
| Salmonella enterica |
12 |
10.4 |
9.1 |
8.7 |
8.8 |
| Lactobacillus fermentum |
12 |
18.4 |
16.1 |
21.6 |
21.9 |
| Enterococcus faecalis |
12 |
9.9 |
8.7 |
14.6 |
14.6 |
| Staphylococcus aureus |
12 |
15.5 |
13.6 |
15.2 |
15.3 |
| Listeria monocytogenes |
12 |
14.1 |
12.4 |
13.9 |
13.9 |
| Bacillus subtilis |
12 |
17.4 |
15.3 |
10.3 |
10.3 |
| Saccharomyces cerevisiae |
2 |
NA |
9.3 |
0.57 |
0.29 |
| Cryptococcus neoformans |
2 |
NA |
3.3 |
0.37 |
0.18 |
Methods
DNA libraries were prepared with two magnetic beads, MAGFLO NGS and gold standard, and processed in parallel. Amplicon sequencing was performed on a MiSeq platform 2x250 bp v2 (Illumina) using the Nextera protocol (Illumina), generating 60 k passed filter reads per sample on average. Fragment analysis was performed using a Fragment Analyzer system (Agilent). Microsynth AG performed experimental procedures and subsequent bioinformatics analyses.
Results
Number of sequencing reads
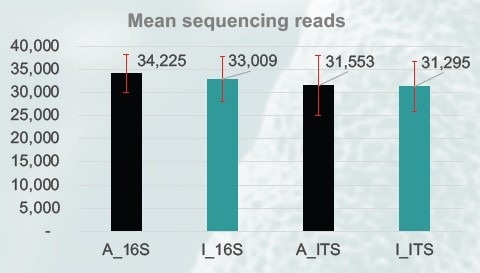
Figure 2. The mean number of sequencing reads obtained for both 16S (V3-V4) and ITS (ITS2) amplicons (n=3). (A: gold standard beads; I: MAGFLO NGS beads). Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Quality of sequencing reads
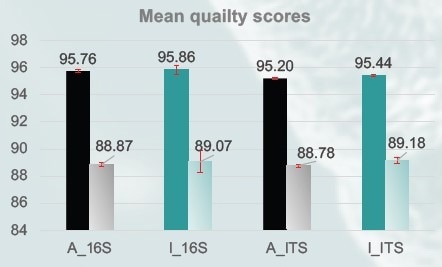
Figure 3. The mean sequencing quality scores for reads with Q20 (filled bars) was around 95%, and with Q30 (shaded bars) around 88%, for both amplicons (n=3). Q values for individual samples were calculated based on mean quality score values from forward and reverse reads. (A: gold standard beads; I: MAGFLO NGS beads). Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Sequencing libraries
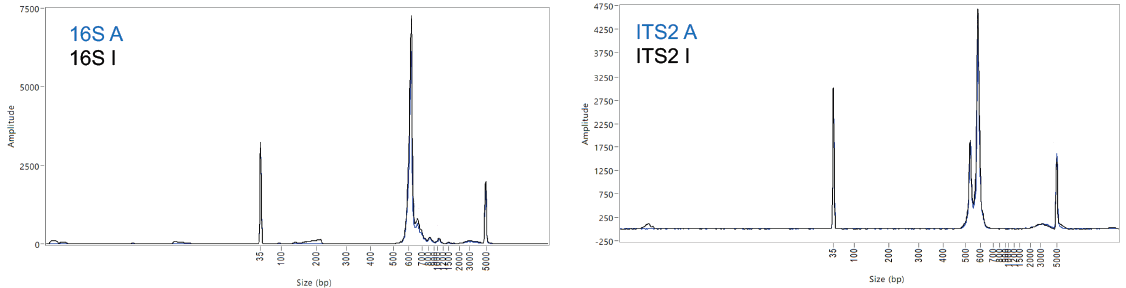
Figure 4. Overlapping DNA fragment profiles obtained prior to sequencing for 16S (left) and ITS2 amplicon (right, 2 visible PCR products accommodate for genetic variants captured with the primers used). (A: gold standard beads; I: MAGFLO NGS beads). Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Theoretical vs. detected microbial species
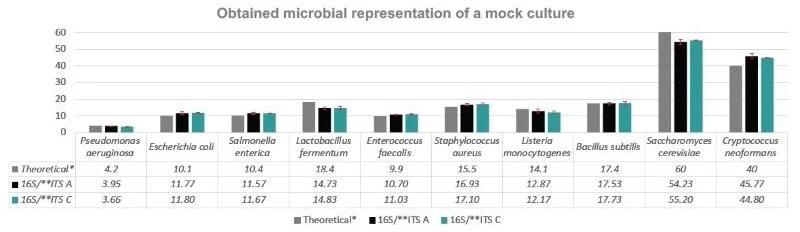
Figure 5. Species aggregation analysis of obtained sequences showed good correlation with theoretical microbial composition of the reference samples. * Theoretical composition is explained in Table 1; ** For ITS, the theoretical composition was based on the copy genome ratio of 0.57/0.37=1.5, therefore 60 and 40% were used, as only 2 species out of 10 were the target for the ITS2-specific primers. Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Alpha diversity of the amplicon
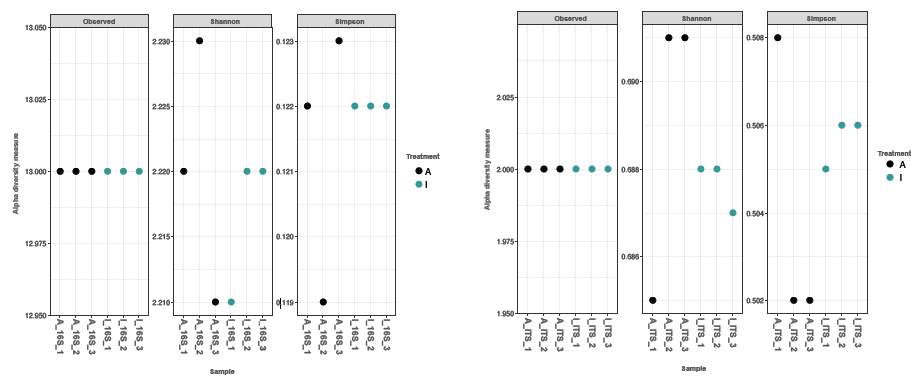
Figure 5. Species aggregation analysis of obtained sequences showed good correlation with theoretical microbial composition of the reference samples. * Theoretical composition is explained in Table 1; ** For ITS, the theoretical composition was based on the copy genome ratio of 0.57/0.37=1.5, therefore 60 and 40% were used, as only 2 species out of 10 were the target for the ITS2-specific primers. Image Credit: Image courtesy of Dagmara Wanda Lewandowska et al., in partnership with ELRIG (UK) Ltd.
Discussion
Sequencing reads with good quality scores ensured reproducible results across triplicates, regardless of the brand of magnetic beads used (Figures 2 and 3). We showed that bead clean-up efficiencies and final DNA fragment lengths were similar for both bead types (Figure 4). The microbial diversity detected correlated with all species' theoretical reference sample composition (Figure 5), and alpha diversity analysis demonstrated no sequencing bias (Figure 6).
Conclusion
The library quality control data and bioinformatics analysis demonstrated the interchangeability of MAGFLO NGS beads and gold standard magnetic beads in a complex NGS amplicon sequencing workflow, particularly in the context of sequencing 16S rRNA and ITS2 phylogenetic markers, ensuring robust microbial composition analysis. The lower reagent price of MAGFLO NGS magnetic beads reduced the processing costs for library preparation, establishing them as a cost-effective alternative to gold standard beads without compromising quality.
References
- Klindworth, A., et al. (2012). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, [online] 41(1), pp.e1–e1. https://doi.org/10.1093/nar/gks808.
- White, T.J., et al. (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. [online] 31, pp.315–322. Available at: https://www.researchgate.net/publication/262687766_Amplification_and_Direct_Sequencing_of_Fungal_Ribosomal_RNA_Genes_for_Phylogenetics [Accessed 23 Oct. 2024].
- https://files.zymoresearch.com/protocols/_d6305_d6306_zymobiomics_microbial_community_ dna_standard.pdf
About INTEGRA Biosciences
 INTEGRA provides innovative solutions for Liquid Handling and Media Preparation applications which serve the needs of their customers in research, diagnostics and quality control laboratories.
INTEGRA provides innovative solutions for Liquid Handling and Media Preparation applications which serve the needs of their customers in research, diagnostics and quality control laboratories.
Their instruments and plastic consumables are developed and manufactured in Zizers, Switzerland and Hudson, NH USA. In order to remain close to their customers, they maintain a direct sales and support organization in North America, the UK, France and Germany, as well as a network of over 100 highly trained distribution partners worldwide.
In recent years they have focused on developing a new and technologically advanced range of handheld electronic pipettes which are simple to use and meet the ergonomic needs of their customers. Today they are proud to offer the widest range of electronic pipettes in the market spanning a range from single channel pipettes up to 384 channel bench-top instruments.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024