A new study by researchers at Germany's Jülich Research Centre reports the inhibition of the aggregation and activation of the spike receptor protein of the virus causing the current pandemic, by a peptide on the angiotensin-converting enzyme 2 (ACE2) receptor. The research is published on the preprint server bioRxiv* in June 2020 and submitted to the Biophysical Journal for peer review.
The ongoing pandemic began in China’s Wuhan, with a cluster of infections caused by a novel coronavirus. This was later identified and named SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), and the syndrome was called coronavirus disease 2019 (COVID-19). The disease quickly spread over the world, with the epicenter shifting from China to South-East Asia and then to Europe, and finally to the US.
NIAIDFollow Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a VERO E6 cell (tan) exhibiting elongated cell projections and signs of apoptosis, after infection with SARS-COV-2 virus particles (green), which were isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Meanwhile, outbreaks in India, Russia, and Brazil continue mostly unchecked, with the global death count now over 400,000 and the number of cases nearing 7 million and growing. The impact of the pandemic has been felt by a heavy toll on human life and health, as well as by the economic disruption in nearly every country in the world.
It is no surprise, therefore, that scientists should be racing to find a vaccine or antiviral drug. One such vaccine target is the spike (S) protein of the coronavirus since this facilitates viral entry into the cell. The S protein is present in the form of trimers, with two subunits, S1 and S2, comprising each trimer.
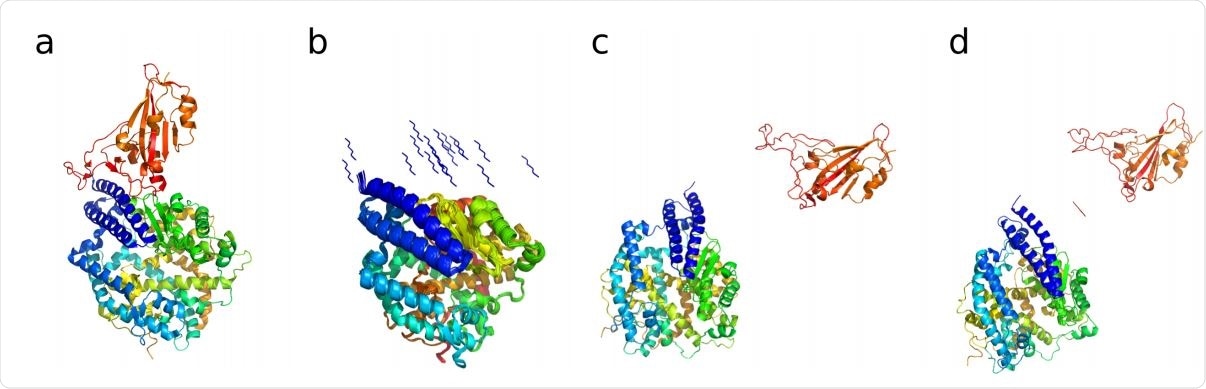
(a) Crystal structure of the SARS CoV-2 spike receptor domain bound with ACE2 (PDB : 6M0J (19)). ACE2 shown in blue, cyan, yellow and green color, the Spike receptor domain is shown in orange. (b) 20 different hexapeptide (YKYRYL) conformations used as initial starting models of independent 50 ns equilibrium MD simulations in explicit solvent. (c) Starting structure of the enhanced aggregation simulation of the spike receptor domain to ACE2. (d) Starting structure of the enhanced aggregation simulation of the same aggregate in the presence of the hexapeptide.
How Does Viral Entry Take Place?
The S1 subunit of the S protein is involved in virus attachment at the cell surface, while the S2 subunit takes part in membrane fusion. Thus, the virus requires the ACE2 molecule on the cell surface as the entry receptor, to enable membrane fusion. At the same time, the host TMPRSS2, a transmembrane serine protease, is involved in priming the S protein before this can happen.
SARS-CoV-2 can make the jump between species to infect humans easily because the ACE2 receptor is highly conserved between different mammalian species. The S2 subunit of the S protein has a receptor-binding domain (RBD) that fits the ACE2 molecule.
The RBD has 5 beta helices running in antiparallel, while alpha-helical and loop motifs connect the beta-sheets. The receptor-binding motif (RBM) is an extended insertion between a pair of beta-sheets, and this binds to the N-terminal end of the ACE2 molecule.
One effective mechanism of preventing viral infection and thus containing the outbreak could be by the inhibition of the ACE2-S protein interaction that leads to viral entry. In the current study, a hexapeptide ((438)YKYRYL(443)) that belongs to the receptor domain of the virus was found to inhibit this interaction efficiently.
The Inhibitory Hexapeptide
In vitro experiments have shown that this peptide prevented the infection of cell cultures by the earlier SARS-CoV. Additionally, it has been found that this peptide also prevents another coronavirus, CoV-NL63, from replicating. Interestingly, this protein fragment has a high affinity for ACE2, being the location of the dominant binding epitope. Its mode of attachment is well established by various imaging techniques so that it has the potential to be used as a template for entry inhibitors against SARS-CoV and similar viruses.
The researchers looked into the ACE2-peptide interaction in greater detail using a technique called molecular dynamics (MD) to provide powerful simulations. This is, therefore, a useful in silico tool that adds to the knowledge provided by experimental studies.
Uncovering the Hexapeptide Binding Mechanics
The aims of the study were, firstly, to quantify the binding affinity of the S protein to the ACE2 molecule. Secondly, the researchers used MD to assess the free energy released when the ligand adsorbs to its binding site at the ACE2 receptor. Thirdly, they looked into how the peptide affects the S protein-ACE2 interaction.
The study shows that the hexapeptide has a high binding affinity to the receptor at 3 clusters at the N-terminal region. According to X-ray studies, these clusters are located at the interface of the spike protein and the receptor. With hexapeptide binding, the spike receptor protein relaxed into an energy minimum that was quite different from the X-ray structure.
The S protein now rotates because of electrostatic repulsion, following which it binds to the ACE2 molecule at a hydrophobic patch rather than via the RBM as suggested by experiments. This is due to the energy minimum.
The simulations show that when the hexapeptide is present, it binds to the S-protein near the binding site to the ACE2 molecule. Following this binding, the S protein changes its pattern of aggregation to the receptor.
The hexapeptide first binds to the spike receptor protein, with a secondary contact at another location. The hexapeptide then diffuses along the surface of the spike receptor domain until it finally binds to the spike protein strongly. The RBM rotates away from the surface of the ACE2 because of electrostatic interactions, but the binding of the RBD to the N-terminal helix, 4 nm farther from the conformation that occurs when the hexapeptide is not present. This very weak binding shows that it is in an inhibited state.
The Effect of the Hexapeptide
In other words, the hexapeptide inhibits the activation of ACE2 and thus prevents membrane fusion and viral entry. Thus both the simulation and the experimental study show that this hexapeptide could be a new mode of treatment to prevent viral entry into human host cells. If the peptide is cleaved by proteases, this dampens the inhibitory effect of the former. Thus, to increase its efficiency, the peptide might need to be chemically modified to prevent such cleavage.
The hexapeptide is also highly specific for the S protein and the ACE2 molecule. Since this rules out another function for this peptide in the human body, it can be considered for development as a potential drug.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources