Eight months into the COVID-19 pandemic, there is still no fully protective therapy against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has infected millions worldwide. Vaccines are being worked on in a host of institutions and countries, mostly targeting the spike protein of the virus. However, the nucleocapsid (N) protein is also extremely abundant and, therefore, a useful target for both vaccine and diagnostic test development. A new study published on the preprint server bioRxiv* in August 2020 reveals the structure of this protein, a valuable step in using it for these purposes.
The N protein has been found to have a mass of 45 to 60 kDa, which suggests that post-translational modifications (PTMs) are taking place. These need to be clearly understood to generate effective vaccines, especially since there is currently a controversy over whether the N protein is glycosylated at all. Uncovering the detailed structure of the N protein may help understand how glycans contribute to the pathogenicity of the virus and vaccine efficacy.
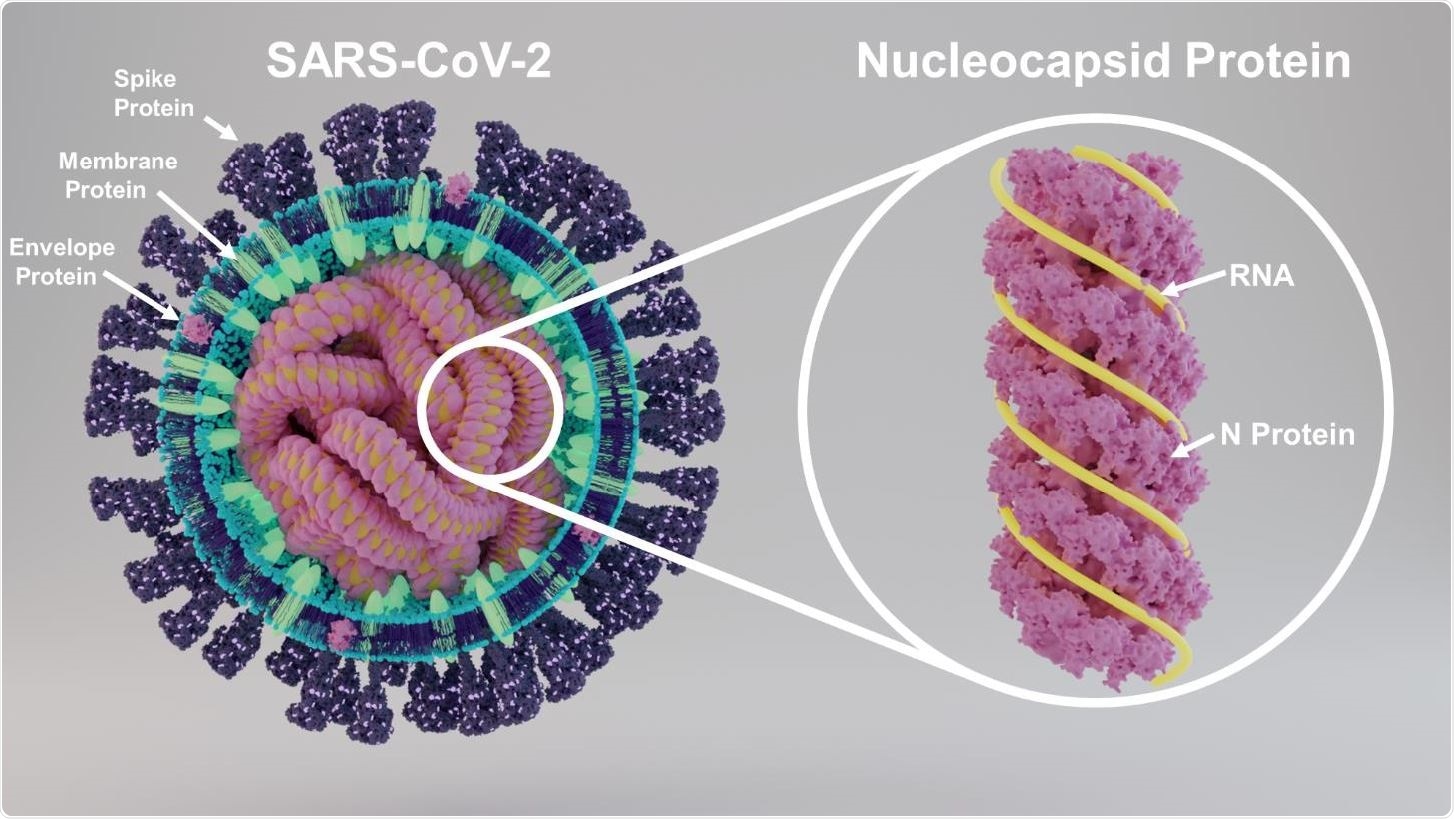
Structure of SARS-CoV-2 showing key proteins and structure of nucleocapsid protein

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Three Domains and What They Do
The N protein contains three highly conserved domains, including an N-terminal RNA-binding domain (NTD) that has electrostatic interactions with the 3’-end of the viral RNA genome through a 55-residue sequence; a C-terminal dimerization domain (CTD), and a central Ser/Arg (SR)-rich linker domain in the middle of this sequence. The NTD binds to RNA, while the CTD mediates oligomerization. The SR-rich linker is mostly responsible for phosphorylation. It also enables molecular movements so that the N protein can interact with other cell components.
The N protein interacts with RNA molecules during the life cycle of the virus within the host cell, including viral replication, genome condensation, and packaging. Such interactions result in the formation of long helices of ribonucleoprotein (RNP), which may compose the external helical part of the nucleocapsid. The internal spherical/ icosahedral core is made up of the N protein, RNA, and the dimerization domain of the M protein.
At the C-terminal end, it interacts with the membrane (M) protein of which the shell is composed to form the genome capsid as virions bud out. Thus, the N protein takes part in the formation of the CoV structure via multiple interactions, while also regulating multiple viral functions like transcription, replication, and modulation of host cell responses.
Inhibiting Antiviral Defenses
Earlier MERS and SARS CoVs had N proteins, which opposed the action of interferons (IFNs), and this is reflected in preliminary results from the study of the SARS-CoV-2 N protein as well. IFNs are cytokines that contribute to immune signaling, especially IFN types I and III, which initiate antiviral defenses in the host cell. The N protein thus modulates the innate immune response of the host and reduces the antiviral response, predisposing to severe infection. The researchers cite recent research: “A recent study found that deceased patient had stronger antibody responses towards N protein while survivors had much more stronger antibody response to S protein highlighting the importance of N protein in disease outcome.”
Diagnostic Potential
Again, recent studies have shown that the N protein can be detected in gargle specimens and nasopharyngeal swabs. This could provide a starting point for rapid and large-scale COVID-19 testing, using tandem mass spectrometry rather than polymerase chain reaction (PCR) and antibody-based tests.
Immunogenic Potential
Thirdly, studies on specimens retrieved from SARS-CoV-2 patients show that the majority of the 26 antigenic sites targeted by specific T cells are not derived from the spike (S) protein. In fact, six of these immunodominant epitopes are from the N protein, making it a potential vaccine candidate that can induce CD8 T cell responses directed explicitly against both spike and other viral proteins.
Identifying Glycan Residues
Early studies show that the N protein is phosphorylated, in fact, the only such SARS-CoV-2 protein and undergoes conformational changes secondary to phosphorylation. However, its glycosylation status is not clear. Some research using amino acid sequencing and software-based glycan detection suggests that N- and O-glycans are likely to be present. The current study is focused on using mass spectrometric techniques to identify the PTMs in the N protein.
The researchers carried out detailed studies of the glycan and glycoprotein residues decorating this protein using a recombinant N protein expressed in cell culture. They found that it has a Ser/Arg-rich (S/R-rich) region, which may contain multiple phosphorylation sites. Phosphorylation may contribute to the regulation of the function of the N protein and its interactions with the M proteins. Further, they detected multiple glycosylation sites and evaluated glycan occupancy.
N- and O-Glycan Binding
There were five potential N-glycosylation sites on the N protein, of which N-glycan binding with distinctive profiles was detected on two sites, as well as one phosphorylation site. One of the N-glycosylation sites had occupancy of above 50%. The NTD has one N-glycosylation and three O-glycosylation sites, according to the current results, which may be regulators of RNA binding.
In the CTD, the researchers observed small amounts of O-glycosylation at two sites and N-glycosylation at one site. Their role in dimerization and self-association of the protein should be urgently explored, say the researchers. Overall, they found multiple O-glycan residues attached to seven sites, as well as less abundant O-glycans on four other sites.
In addition, they investigated the occupancy of the glycosylation sites, as well as differences in the nature and occupancy of these sites in different variants of the protein. They looked at sialic acid and fucose linkages.
Glycan Attributes
Most glycans were high-mannose and complex types of glycans, making up 73% of the total, with hybrid N-glycans forming a little more than a fifth. They also found N-glycans to be present at 5% of the total. The O-glycans they identified on the N protein belonged to four types, from Core 1 to Core 4, but 96% belonged to the first two types. Some uncommon N-glycans with unusual linkages were observed. This could indicate, as in other biological pathways, that these glycans are playing essential roles and are key to successful viral infection. These carbohydrates can be targeted by immune recognition cells as well from both the innate and adaptive pathways.
The researchers predict that these findings on O- and N-glycosylation of the viral N protein “could be a key aspect in the development of therapeutic agents that specifically and efficiently block the coronavirus replication, transcription, and viral assembly.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.