A startling new study reports the rapid rise to dominance of a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain in Italy. This strain, termed the GV clade, is characterized by a single point mutation.
The study's findings emphasize the importance of understanding the virus's mutation dynamics. These alterations in the key antigens could affect accurate diagnoses and undermine the efficacy of vaccines and other interventions.
Epidemiological studies are key to evolving a winning set of strategies against any infectious disease. The current pandemic of COVID-19 is no exception. Multivalent therapeutics and prophylactics will be essential long-term to prevent recurring waves of COVID-19 cases.
Scientists first reported a new SARS-CoV-2 variant with the A222V substitution in the spike glycoprotein in Spain during March, 2020. This spread quickly across the European continent from June onwards, and became the GV clade's ancestor. About 7% of current genomes uploaded to the GISAID database are due to this virus clade at present.
Study details
The current study is based on regular whole genome sequence (WGS) analyses carried out by the researchers as part of the COVID-19 surveillance program in the Lazio region of Italy.
The investigators aimed to understand the place held by this strain in the new wave of infections currently hitting Italy. In doing this, they explored the genome sequences of samples from June to October, 2020, in their laboratory at the National Institute for Infectious Diseases (INMI) in Rome. In total, there were 57 whole genome sequences. Of these, 33 were from the INMI laboratory, and 24 from others in Italy.
Among them, 46 sequences were obtained during surveillance. Another 5 sequences were from individuals with a history of travel, taken in the second half of August, when the second wave may be said to have begun in Italy, and 6 from the third week in October, which represents variants in current circulation. The analysis also included other sequences from Italy collected during this period and uploaded by other researchers to GISAID; these totaled 89 sequences.
The clustering analysis provided 57 unique sequences, which were shown, on phylogenetic analysis, to have one cluster of correlated sequences from clade GR, all from June, and retrieved from Rome, Lazio. There were 10 sequences, 9 from this laboratory and 1 from another laboratory in Sardinia, which was retrieved from GISAID, from August, and contained the A222V substitution (being part of the GV clade). This comprises 17.5% of all Italian sequences.
The infected individuals with these variants were (August-time) travelers returning to Italy from Sardinia or Spain, as well as those (in October) without known travel history.
During the study period, 3 other clades – namely, GR, G and GH – were circulating, with the GR clade comprising the dominant strains at around 68% of isolates. G and GH made up around 9% and 5% of circulating viral strains.
It is noteworthy that there were two other isolates from travelers, collected in August, one from an individual returning from Croatia and one from a Malta returnee, which belonged to GR and G clades, respectively. They also found three other isolates from a Neapolitan laboratory, which was from clade G as well.
Though this study depends on a small number of sequences, the findings show a sudden rise in the number of GV clade variants in circulation in this country, such that the A222V variant now makes up over 11% of sequences in the period from June to October. The earliest GV isolates were observed in August (on the 25th) and came from a Sardinian subject, with two others from travelers returning from that island or from Spain. However, all GV sequences in samples collected in October were from patients without a known history of travel, and presumably represent community transmission.
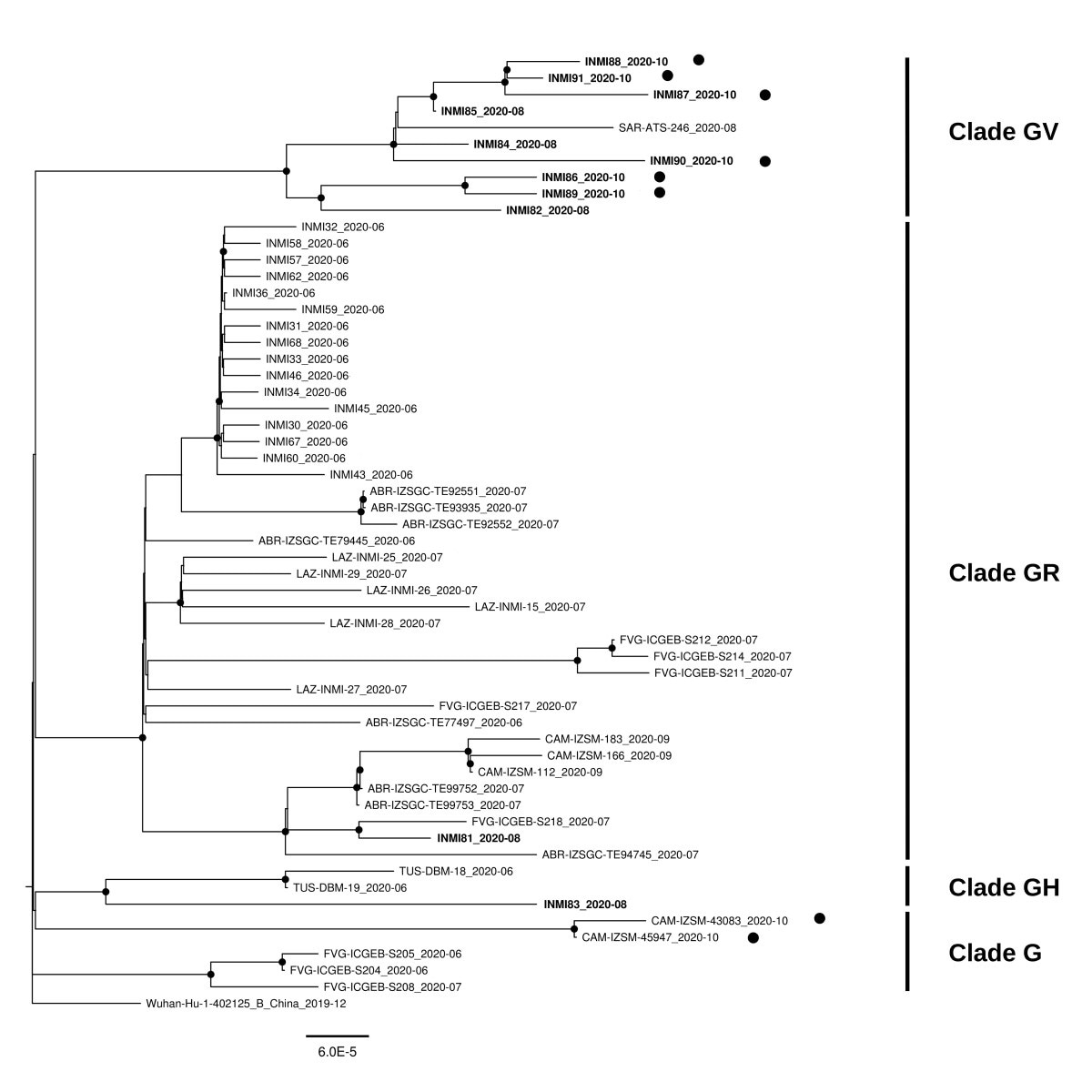
Maximum likelihood phylogenetic tree of all SASR-CoV-2 unique sequences obtained in Italy in from 1st June and 2nd November 2020 (n=57), including 33 sequences obtained in our laboratory, at the National Institute for Infectious Diseases (INMI) in Rome, Italy, and 24 sequences from other Institutions in Italy. Sequences obtained at INMI are indicated by the name that also includes the collection date. To underline the chronological order of sequences obtained in our laboratory, sequences obtained from August onward are highlighted in bold. Italian sequences obtained in late October are highlighted with a bullet. Clades are indicated on the side of the tree, following the GISAID nomenclature. All nodes with bootstrap values higher than 75 are highlighted with a black point. Maximum likelihood phylogenetic tree of all SASR-CoV-2 unique sequences obtained in Italy in from 1st June and 2nd November 2020 (n=57), including 33 sequences obtained in our laboratory, at the National Institute for Infectious Diseases (INMI) in Rome, Italy, and 24 sequences from other Institutions in Italy. Sequences obtained at INMI are indicated by the name that also includes the collection date. To underline the chronological order of sequences obtained in our laboratory, sequences obtained from August onward are highlighted in bold. Italian sequences obtained in late October are highlighted with a bullet. Clades are indicated on the side of the tree, following the GISAID nomenclature. All nodes with bootstrap values higher than 75 are highlighted with a black point. Maximum likelihood phylogenetic tree of all SASR-CoV-2 unique sequences obtained in Italy in from 1st June and 2nd November 2020 (n=57), including 33 sequences obtained in our laboratory, at the National Institute for Infectious Diseases (INMI) in Rome, Italy, and 24 sequences from other Institutions in Italy. Sequences obtained at INMI are indicated by the name that also includes the collection date. To underline the chronological order of sequences obtained in our laboratory, sequences obtained from August onward are highlighted in bold. Italian sequences obtained in late October are highlighted with a bullet. Clades are indicated on the side of the tree, following the GISAID nomenclature. All nodes with bootstrap values higher than 75 are highlighted with a black point.
What are the Implications?
In short, the research suggests that the GV clade was introduced into Italy in August, perhaps first in Sardinia and Lazio, among other regions, by travelers re-entering the country, and then spread widely among the local inhabitants. Such community transmission is still continuing to take place. More studies are required to understand the occurrence of this clade in other regions of Italy, and how it affects the spread of this virus.
The A222V mutation is in a spike protein region that does not impinge on the receptor-binding domain. Nevertheless, it is important to evaluate the mutation's role in enhancing the efficiency of viral transmission. Again, further studies should focus on the location of the mutation in a possible B cell-binding site, which may mediate altered immune cell recognition.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bartolini, B. et al. (2020). The newly introduced SARS-CoV-2 variant A222V is rapidly spreading in Lazio region, Italy. medRxiv preprint. doi: https://doi.org/10.1101/2020.11.28.20237016,
https://www.medrxiv.org/content/10.1101/2020.11.28.20237016v1
- Peer reviewed and published scientific report.
Bartolini, Barbara, Martina Rueca, Cesare Ernesto Maria Gruber, Francesco Messina, Fabrizio Carletti, Emanuela Giombini, Eleonora Lalle, et al. 2020. “SARS-CoV-2 Phylogenetic Analysis, Lazio Region, Italy, February–March 2020.” Emerging Infectious Diseases 26 (8): 1842–45. https://doi.org/10.3201/eid2608.201525. https://wwwnc.cdc.gov/eid/article/26/8/20-1525_article.