Coronaviruses are a large family of viruses that can infect both animals and humans. First identified in the 1960s, the viruses cause respiratory illness and are well-known for their surface's crown-like spikes.
A new study by researchers at the University of the Western Cape aimed to determine why some coronaviruses are more virulent than others. The team speculates that the increased flexibility of the PDZ-binding motif (PBM) of SARS-CoV, MERS-CoV, and SARS22 CoV-2 to be more flexible than the PBM of hCoVs 229E, NL63, OC43, and HKU1.
Published on the preprint serve bioRxiv*, the study highlights the differences between the coronaviruses based on the coronavirus envelope, a crucial contributor to severe human coronaviruses' pathogenesis. These differences may also shed light on possible drug or vaccine development.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Study background
The disease severity and immunopathology of SARS-CoV and SARS-CoV-2 infections have been attributed to the envelope protein. However, there have been no studies showing whether this is also the case for MERS-CoV-2 infections.
Studying the envelope protein in each of the human coronaviruses can help determine if it impacts viral virulence.
The SARS-CoV-2 E protein has been known to interact with host cell proteins syntenin and the protein tied to Caenorhabditis elegans lin-7 protein 1 (PALS1) cytoplasm of host cells, highlighting the importance of its topology.
Meanwhile, the topology of the E protein for the less virulent human coronaviruses is unknown. No evidence shows if it adopts a topology similar to the more virulent coronaviruses.
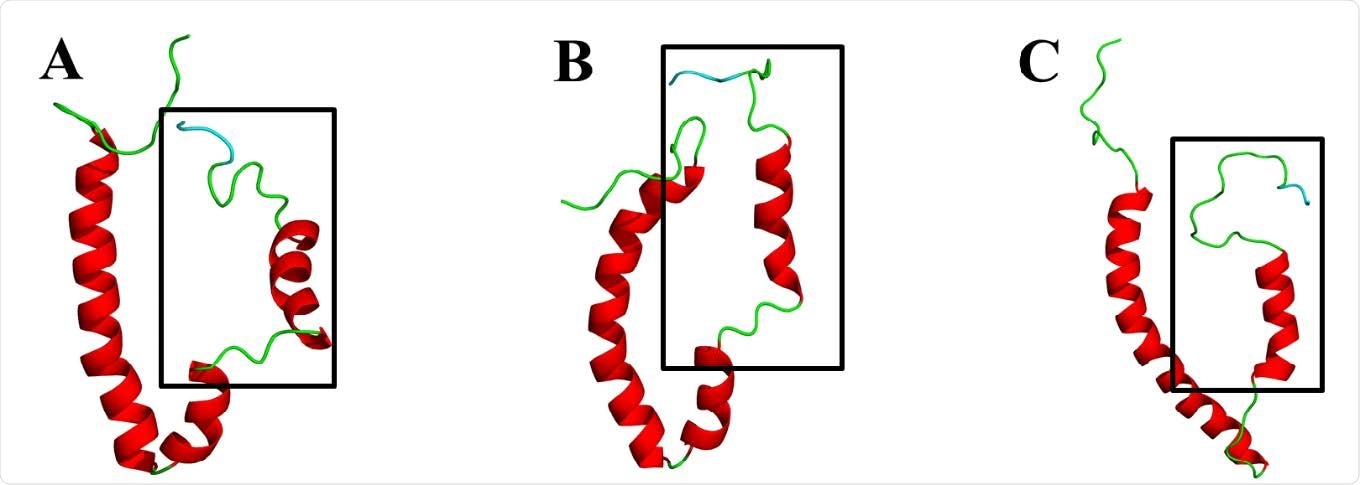
Cartoon representation of the three-dimensional (3D) models of the envelope (E) protein of the more virulent hCoVs. A. SARS-CoV E. B. SARS-CoV-2 E. C. MERS-CoV E. Models were generated using MODELLER software and based on two nuclear magnetic resonance (NMR)-resolved structures for SARS-CoV E (PDBID: 5x29 and PDBID: 2mm4) obtained from the protein data bank (PDB) (Eswar, et al., 2008; Li, et al., 2014; Sali and Blundell, 1993; Surya, et al., 2018). Models of the SARS-CoV and SARS-CoV-2 E proteins were generated from the 5x29 template, whereas the MERS-CoV E protein was generated from 2mm4 template. The PDZ-binding motif (PBM) is colored in cyan and the squared region shows the variable C-terminal domain.
The study
The complexity of membrane proteins presences a challenge in studying their nature experimentally. When it is impossible to conduct experiments, bioinformatics software tools can be used to dissect topologies.
The researchers used topology prediction programs and 3D modeling software to predict and envision structural aspects of the E protein related to its functions in the current study.
The team studied all seven hCoV E proteins, and they found that these viruses adopted different topologies, with distinctions between the more virulent and less virulent ones. The study findings showed that the PBM of SARS-CoV, MERS-CoV, and the SARS-CoV-2 are more flexible than the PBM of OC43, HKU1, and 229E.
The study showed that the more flexible the PBM is, the more virulent the virus is. Increased flexibility may provide the more virulent viruses more freedom, allowing them to bind to various host proteins. This can contribute to a more severe form of infection.
"This is the first paper to predict the topologies and model 3D structures of all seven hCoVs E proteins, providing novel insights for possible drug and vaccine development," the researchers noted in the paper.
Though much is still unclear about the hCoV E protein's topology, the study provided the first step for further studies. The study was the first to predict and generate 3D models of the E protein for all seven coronaviruses.
The study findings may provide a clear explanation of the differences in virulence of the viruses.
COVID-19 toll
Finding ways to find effective therapies for COVID-19 can help stem the pandemic. The ongoing global health crisis has now caused over 117 million infections globally. More than 2.61 million people have died due to the virus.
The United States reports the highest number of cases, reaching over 29 million, with over 527,000 deaths. India and Brazil also report high number of cases, topping 11.24 million and 11.12 million cases.
Since December 2020, many countries have started vaccinating residents, focusing first on healthcare workers, front-line workers, and the elderly.
With the surging number of cases, scientists believe finding ways to produce therapies and vaccines effectively is crucial to combat the pandemic.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Source:
Journal reference:
- Preliminary scientific report.
Schoeman, D., Cloete, R., and Fielding, B. (2021). Comparative studies of the seven human coronavirus envelope proteins using topology prediction and molecular modelling to understand their pathogenicity. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.03.08.434384v1