A team of scientists from the USA and Canada has recently characterized the antibodies developed against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in response to an mRNA-based coronavirus disease 2019 (COVID-19) vaccine. The findings reveal that although the vaccine is capable of inducing a robust polyclonal antibody response, the majority of the antibodies are devoid of virus-neutralizing activity. The study is currently available on the medRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Since the development of potential vaccines against SARS-CoV-2, many preclinical and clinical studies have been done to evaluate the innate and adaptive immune responses developed by the host immune system in response to vaccine antigens. Some of these vaccines have recently received emergency use approval and are currently rolling out in many countries. Because of its direct involvement in mediating viral entry into host cells via angiotensin-converting enzyme 2 (ACE2) receptor, the spike receptor-binding domain (RBD) of SARS-CoV-2 is regarded as the most potent target for developing COVID-19 vaccines. Studies investing in vaccine-induced immune responses have shown that monoclonal antibodies that bind spike RBD can potentially block the virus-host interaction, and thus, can effectively neutralize the virus. However, a growing-pool of recent evidence suggests that apart from spike RBD or full-length spike protein, the N-terminal domain (NTD) of the spike S1 subunit contains neutralizing epitopes, and thus, can serve as potential targets for anti- SARS-CoV-2 antibodies.
In the current study, the scientists have profiled the polyclonal antibodies and plasmablast-derived monoclonal antibodies developed against SARS-CoV-2 in response to an mRNA-based, spike-targeting COVID-19 vaccine produced by Pfizer-BioNTech.
Study design
In the study, six adult participants received two doses of the mRNA-based COVID-19 vaccine. Blood samples were obtained from the participants before vaccination and after the 1st and 2nd doses of vaccination. The kinetics of vaccine-induced anti-RBD and anti-NTD antibodies were compared with the antibody kinetics of 30 COVID-19 survivors who have developed a wide-range of anti-SARS-CoV-2 antibodies in response to natural infection.
Important observations
The analysis of antibody kinetics revealed that the levels of anti-spike and anti-spike RBD polyclonal antibodies were significantly higher in vaccinated individuals than that in COVID-19 survivors. Among vaccinated individuals, the antibody response reached a peak value one week after the 2nd dose, followed by a gradual decline with time. Interestingly, COVID-19 survivors with low antibody titers exhibited a higher ratio of binding to neutralizing antibodies than those with high titers. However, at the time of peak response, the vaccinated individuals showed the highest ratio of binding to neutralizing antibodies, which indicates that the majority of vaccine-induced antibodies are non-neutralizing in nature.
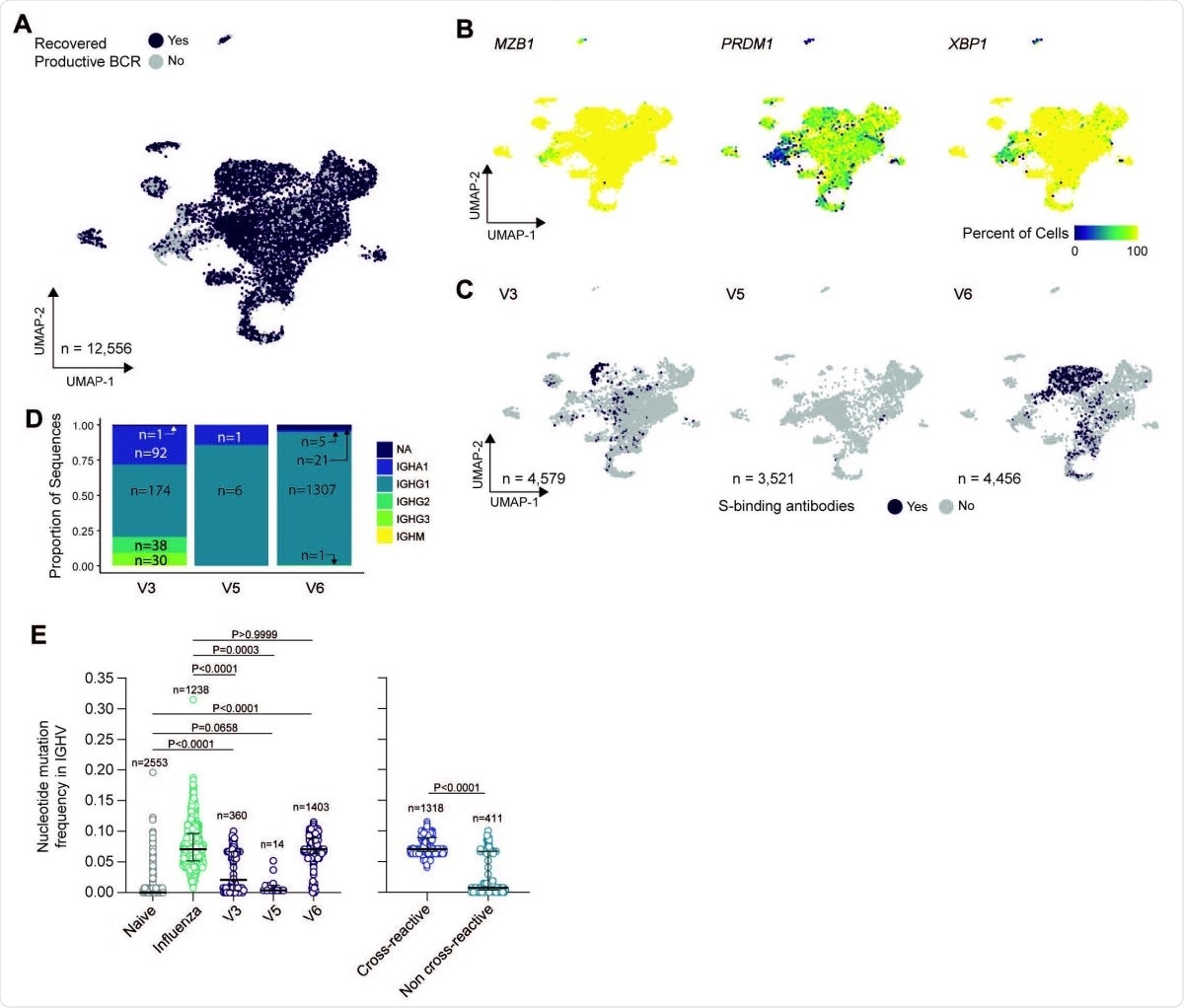
Characterization of bulk sorted plasmablasts via single-cell RNA sequencing. (A) Uniform manifold approximation and projection (UMAP) of scRNAseq from bulk plasmablast with recovered BCR sequences (purple) or unrecovered (grey). (B) UMAP overlay of percent of cellular population expressing MZB1, PRDM1, and XPB1. Hexbin equals 80 individual cells. (C) UMAP overlay of BCR sequences with confirmed spike binding activity. (D) Proportional composition of heavy chains genes in the spike binding sequences broken down by sample. (E) Comparison of nucleotide-level mutation frequency in immunoglobulin heavy chain variable (IGHV) genes between plasmablasts clonally related to spike binding mAbs from SARS-CoV-2 vaccinees, plasmablasts sorted from PBMCs one week after seasonal influenza vaccination and found in vaccine-responding B cell clones, and naïve B cells found in blood of an influenza vaccinee (left panel); and between plasmablasts from SARS-CoV-2 vaccinees found to be clonally related to spike-binding mAbs that were, respectively, cross-reactive and non-cross-reactive to human ß- coronaviruses spike proteins (right panel).
Regarding pre-existing immunity against seasonal coronaviruses, the scientists observed that the mRNA vaccine significantly increased the levels of antibodies directed against the spike protein of seasonal β-coronaviruses, such as OC43 and HKU1. However, they did not observe any such effect of the vaccine on α-coronaviruses, such as 229E and NL63.
To characterize vaccine-induced B cell response, the scientists isolated plasmablasts from the blood samples of the vaccinated individuals and subsequently screened all plasmablast-derived monoclonal antibodies. Interestingly, they observed that most antibodies were directed against the spike NTD and that only a small fraction of the antibodies recognized the spike RBD. These observations indicate that the vaccine is capable of generating both anti-RBD and anti-NTD monoclonal antibodies. With further analysis, they observed that the majority of these monoclonal antibodies were unable to neutralize SARS-CoV-2.
The scientists used biolayer interferometry to examine the affinity of different RBD mutation-containing viral variants for human ACE2. They observed that almost all the RBD mutations they analyzed (N501Y, Y453F, N439K, E484K, and K417N), either alone or in combination, significantly increased the viral affinity for ACE2 receptor. Furthermore, they examined the reactivity of serum samples collected from vaccinated individuals and COVID-19 survivors against various RBD mutants. The findings revealed that all polyclonal sera from COVID-19 survivors bound better to N501Y RBD than the wildtype RBD, whereas about 40% reduction in binding was observed for RBD-mutated B.1.351 variant. However, the sera from vaccinated individuals showed the highest reduction of two-fold against RBD mutations present in B.1.351 and P.1 variants.
By analyzing the PV14252 variant of SARS-CoV-2 that contains several NTD mutations and the E484K RBD mutation, the scientists observed that out of 7 plasmablast-derived monoclonal antibodies, only two anti-RBD antibodies neutralized both wildtype SARS-CoV-2 and the PV14252 variant with similar efficiency. In contrast, all five anti-NTD antibodies completely lost their ability to neutralize the PV14252 variant because of the NTD mutations present in that viral variant.
Study significance
The study reveals that the mRNA-based COVID-19 vaccine (Pfizer-BioNTech) induces both anti-RBD and anti-NTD antibodies; however, the majority of these antibodies are non-neutralizing in nature. Moreover, the study highlights the importance of NTD mutations in viral variants that can significantly abolish the virus-neutralizing ability of monoclonal antibodies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Amanat F. 2021. The plasmablast response to SARS-CoV-2 mRNA vaccination is dominated by non-neutralizing antibodies that target both the NTD and the RBD. MedRxiv. doi: https://doi.org/10.1101/2021.03.07.21253098, https://www.medrxiv.org/content/10.1101/2021.03.07.21253098v1
- Peer reviewed and published scientific report.
Amanat, Fatima, Mahima Thapa, Tinting Lei, Shaza M. Sayed Ahmed, Daniel C. Adelsberg, Juan Manuel Carreño, Shirin Strohmeier, et al. 2021. “SARS-CoV-2 MRNA Vaccination Induces Functionally Diverse Antibodies to NTD, RBD, and S2.” Cell 184 (15): 3936-3948.e10. https://doi.org/10.1016/j.cell.2021.06.005. https://www.cell.com/cell/fulltext/S0092-8674(21)00706-6.