A team of scientists from the United States has recently examined the trajectory of early immune biomarkers in response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. They have identified specific plasma proteins that can accurately predict the clinical and virological outcomes in SARS-CoV-2 infected patients. The study is currently available on the medRxiv* preprint server.
Since its emergence in December 2019 in Wuhan, China, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has infected 217 million people and claimed 4.5 million lives globally. Although the disease remains asymptomatic in most patients, it can develop into severe life-threatening consequences in vulnerable people, including older adults and those with comorbidities.
Early immune responses to SARS-CoV-2 infection determine the disease severity and dynamics of virus-specific memory immune responses. Upon infection, recognition of viral RNA by pattern recognition receptors, including toll-like receptors (TLRs) and RIG-I-like receptors (RLRs), results in the production of pro-inflammatory mediators and interferons. The stimulation of interferon-responsive genes by interferons subsequently leads to induction of early antiviral response and initiation of an adaptive immune response. An insufficient interferon response during the early infection phase can increase the risk of severe COVID-19.
In the current study, the scientists have conducted a proteomic and transcriptomic analysis to identify immune markers induced during the early course of SARS-CoV-2 infection. They have utilized these markers to predict viral load, disease severity, and memory immune responses in patients with mild to moderate COVID-19.
Study design
The scientists analyzed longitudinal samples collected from COVID-19 patients enrolled for a randomized clinical trial of Peginterferon Lambda-1a, a type III interferon. In the trial, mild to moderate COVID-19 patients were enrolled within 72 hours of diagnosis and followed up for 7 months post-infection.
They analyzed the samples to identify immune biomarkers induced within the first 2 weeks of symptom onset. Using a machine learning model, they have examined the ability of some early-response plasma proteins to predict viral load, disease severity, and memory T cell and antibody responses.
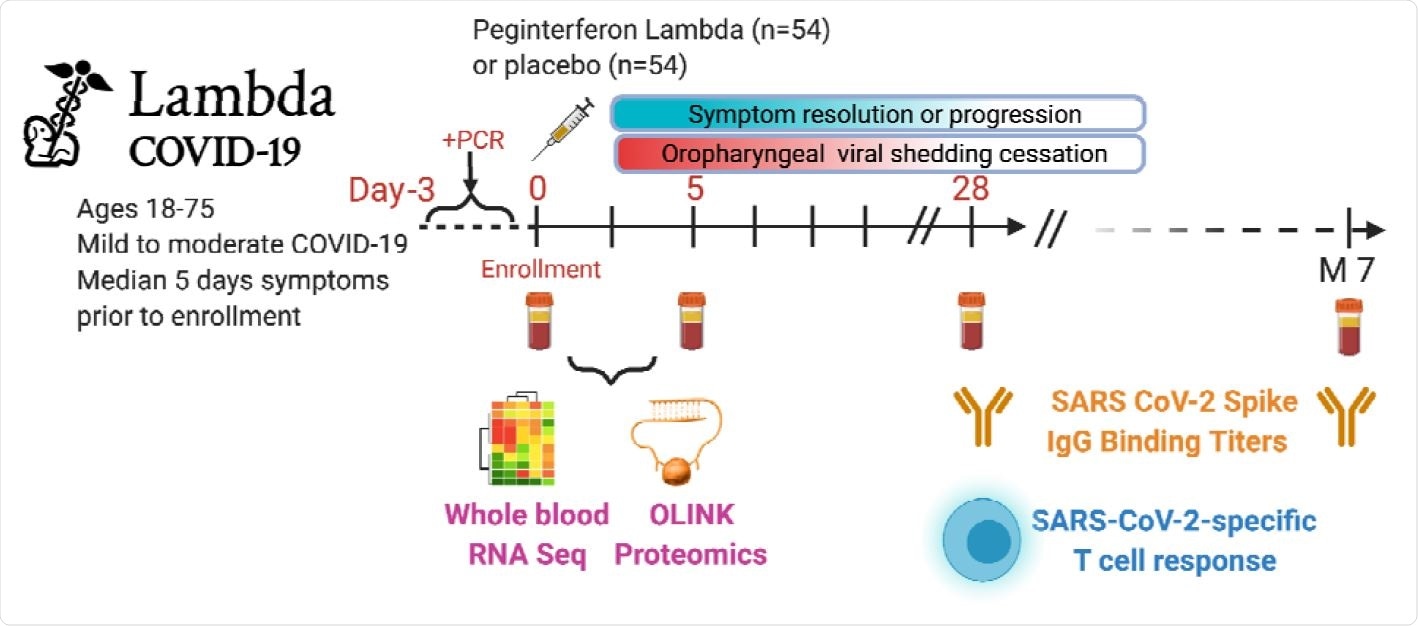
Study schema. Outpatients (n=108) with PCR-confirmed SARS-CoV-2 infection 2 and swab obtained within 72 hours of randomization were enrolled in a Phase 2 clinical trial of subcutaneous Peginterferon lambda vs. placebo. In-person follow-up visits were conducted at day 1, 3, 5, 7, 10, 14, 21, 28,and month 7 post-enrollment, with assessment of symptoms and vitals, and collection of oropharyngeal swabs for SARS-CoV-2 testing. Blood obtained at Day 0 and 5 were evaluated by whole blood transcriptomics (RNA Sequencing), plasma proteomics (Olink), and SARS-CoV-2 specific antibodies. Clinical outcomes assessed included duration of symptoms and duration of virologic shedding. Immunologic outcomes assessed including SARS-CoV-2-specific T cell responses at day 28, and antibody responses at day 28 and month 7. Created with biorender.com.
Important observations
The scientists identified 38 immune pathways and 10 plasma proteins that differed as a function of time since symptom onset. Based on the response pattern, these pathways and proteins were divided into 4 clusters. Cluster 1 contained interferon-related pathways, natural killer cell activation pathways, and interferon-stimulated proteins (MCP-1, MCP-2, CXCL10, and CXCL11). These markers showed a peak value at the time of symptom onset, followed by a gradual decline over time. In cluster 2, which contained Interferon-γ, chemokines CXCL1 and CXCL6, innate immune response pathways, and T cell activation pathways, peak values were observed 1 – 5 days after the symptom onset. The B cell activation pathways identified in cluster 3 showed a peak between 10 and 14 days after symptom onset. The cluster 4 markers, including anti-spike antibodies and B cell differentiation pathways, gradually increased after the symptom onset.
Taken together, these observations indicate that the activation of interferon signaling, B cells, T cells, and natural killer cells and production of anti-SARS-CoV-2 antibodies occur within the first 15 days of symptom onset.
Association between immune responses and disease severity
Of 108 enrolled patients with mild to moderate COVID-19, eight developed serious complications eventually. To identify immune pathways and plasma proteins responsible for severe disease, the scientists compared these patients with those who eventually became virus-free or asymptomatic. They identified 17 immune pathways and 24 plasma proteins that were significantly associated with disease severity. These markers were interferon-induced proteins and natural killer cell activation pathways, indicating the involvement of early immune responses in modulating disease severity.
Association between immune responses and disease outcome
The scientists identified 36 plasma proteins associated with disease outcome, including RIG-I, CCL20, CCL25, Keratin 19, and amphiregulin. Increased expressions of these proteins were negatively associated with viral load. These proteins were also associated with higher levels of spike-specific T cell and antibody responses. Similarly, multiple immune pathways, including complement and B cell activation pathways and antibody response pathways were identified as regulators of viral load.
With further analysis, 4 plasma proteins and 12 immune pathways were identified that showed significant association with time since symptom onset. Specifically, peaked levels of interferon-gamma response, interferon-induced cytokines, and T cell-secreted cytokines were observed within the first 5 days of symptom onset. In addition, B cell activation and antibody production pathways reached peaked levels between 10 and 15 days after symptom onset. Together these findings indicate that induction of an early interferon response and subsequent pro-inflammatory cytokine signaling determine the clinical, virological, and immunological outcomes in COVID-19 patients.
Interestingly, the findings revealed that a high plasma level of RIG-I is associated with less viral load, higher disease severity, and increased T cell and antibody responses. In addition, a strong association was observed between RIG-I and apoptosis (programmed cell death).
Regarding immune responses to COVID-19 vaccines, the findings indicated that immune responses induced by the first dose of Pfizer/BioNTech-developed BNT162b2 vaccine were similar to that induced by natural SARS-CoV-2 infection. However, rapid induction of adaptive immune response and a lack of neutrophil response was observed in response to the second vaccine dose.
Machine learning models
The scientists conducted predictive modeling to identify proteins that could predict viral load, disease severity, and immune responses. They finally selected 8-10 plasma proteins measured at the early infection phase and observed that these proteins could accurately predict all tested outcomes.
Study significance
The study identifies a cytosolic pattern recognition receptor (RIG-I) and pro-inflammatory cytokines (MCP1, 10 MCP2, and MCP3) as potential biomarkers to predict clinical and immunological outcomes in COVID-19 patients. Moreover, the study highlights the importance of machine learning models in identifying high-risk COVID-19 patients using 8 – 10 plasma proteins.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hu Z. 2021. Early immune responses have long-term associations with clinical, virologic, and immunologic outcomes in patients with COVID-19, medRxiv, https://doi.org/10.1101/2021.08.27.21262687, https://www.medrxiv.org/content/10.1101/2021.08.27.21262687v1
- Peer reviewed and published scientific report.
Hu, Zicheng, Kattria van der Ploeg, Saborni Chakraborty, Prabhu S Arunachalam, Diego AM Mori, Karen B Jacobson, Hector Bonilla, et al. 2022. “Early Immune Markers of Clinical, Virological, and Immunological Outcomes in Patients with COVID-19: A Multi-Omics Study.” Edited by Miles P Davenport. ELife 11 (October): e77943. https://doi.org/10.7554/eLife.77943. https://elifesciences.org/articles/77943.