We know little about the origin and reservoir of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the ongoing coronavirus disease 2019 pandemic (COVID-19). However, horseshoe bats tested in Yunnan province, China, have been found to carry the closest relatives of SARS-CoV-2.
Coronaviruses and bats
The origin, reservoir, diversity, and extent of circulation of SARS-CoV-2 ancestors are unknown; however, Horseshoe bats are believed to be the primary natural reservoirs for SARS-related coronaviruses. Horseshoe bats from several provinces in China have been found to carry several coronavirus species. Southeast Asia is home to more than 25% of the world's bat species and is considered a hotspot for emerging zoonotic diseases. A close relative of SARS-CoV-2 was identified in bats captured from a cave in Thailand in June 2020.
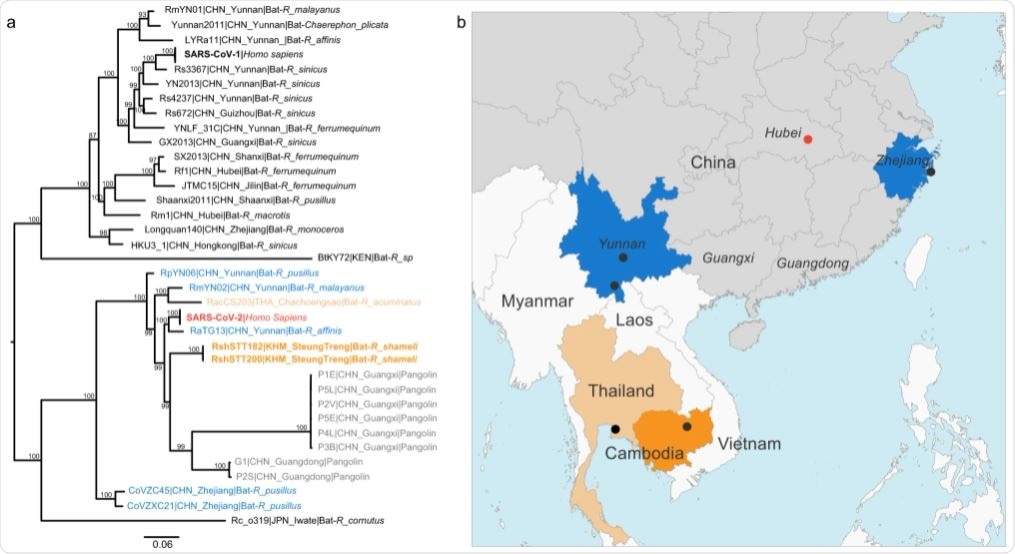
a Maximum likelihood phylogeny of the subgenus Sarbecovirus (genus Betacoronavirus; n = 39) estimated from complete genome sequences using IQ-TREE and 1000 replicates. The coronaviruses of the SARS-CoV-2 lineage are color coded by country of sampling as on the map. In orange, Cambodia, light orange, Thailand and blue, China. Taxa names include the isolate name, country and province of sampling, and host. The scientific names of the hosts are abbreviated as follows: Bats: R. affinis, Rhinolophus affinis; R. sinicus, Rhinolophus sinicus; R. ferrumequinum, Rhinolophus ferrumequinum; R. malayanus, Rhinolophus malayanus; R. acuminatus, Rhinolophus acuminatus; C. plicata, Chaerephon plicata; R. pusillus, Rhinolophus pusillus; R. macrotis, Rhinolophus macrotis; R. monoceros, Rhinolophus monoceros; R. cornutus, Rhinolophus cornutus; Pangolin: M_javanica, Manis javanica and human: H. sapiens, Homo sapiens. A maximum clade credibility tree is available in Supplementary Fig. 3. b map of parts of China and Southeast Asia. Regions where viruses of the SARS-CoV-2 lineage were sampled are colored as in the tree. A black dot indicates a sampling site when known, and the red dot shows the location of Wuhan, where the first cases of SARS-CoV-2 infection were reported.
Cambodia
UNESCO and the National Authority of Preah Vihear mandated the Muséum national d'Histoire naturelle (MNHN, Paris, France) to conduct a mammal survey in northern Cambodia in 2010. During this survey, bats were captured using mist nets and harp traps from two provinces, Preah Vihear and Ratanakiri. Bat diversity on the two sides of the Mekong River was compared.
Likewise, USAID funded the PREDICT project to sample bats to detect and discover viruses with a pandemic and zoonotic potential. Oral and rectal swabs were collected from bats from 2012 to 2018.
The samples were tested for SARS-CoV-2 related viruses. Two bats positive for viruses closely related to SARS-CoV-2 were collected during the MNHN mission.
The investigators tested 430 archived samples, including 162 oral swabs and 268 rectal swabs. Sixteen rectal swabs (3.72%) tested positive for coronaviruses - 11 alphacoronaviruses and five betacoronaviruses. Two of the five betacoronavirus samples further tested positive using a specific reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) test. These two samples were from rectal swabs of Shamel's horseshoe bats sampled in December 2010 in the Steung Treng province in Cambodia. Oral swabs from these bats tested negative for betacoronaviruses.
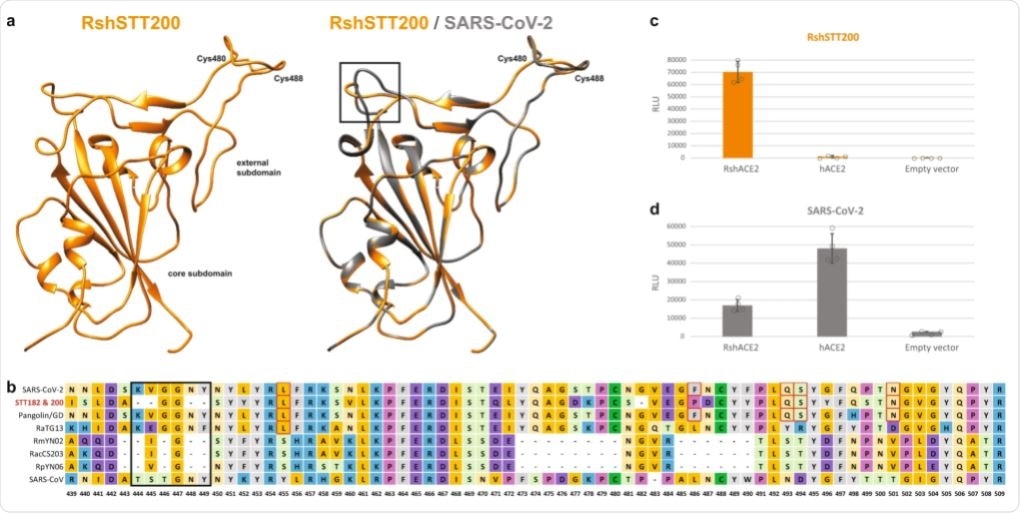
a Homology modeling of the RBD structure. The three-dimensional structure of the RshSTT200 Spike RBD was modeled using the Swiss-Model program employing the structure of SARS-CoV-2 (PDB: 6yla.1) as a template. The core and external subdomains are colored orange, and gray for RshSTT200 and SARS-CoV-2, respectively. The shortening a loop near the receptor-binding site of RshSTT200 are indicated by a black rectangle. The cysteines involved in a conserved disulfide bond are indicated. b Alignment of the receptor-binding motif amino acid sequences of selected betaCoVs. c RshSTT200 spike (deleted for the last 21 amino acids) pseudovirus entry into HEK293T cells transfected with either RshACE2, hACE2 or an empty vector (pLenti-puro). d SARS-CoV-2 spike pseudovirus entry into HEK293T cells transfected with either RshACE2, hACE2 or an empty vector (pLenti-puro). Data are represented as mean ± standard deviation of technical replicates (n = 4) and are representative of three independent experiments. Source data are provided as a Source Data file.
Genetic similarity with SARS-CoV-2
The RNA from the two coronaviruses was then processed for next-generation metagenomic sequencing. The sequenced genome was compared to the SARS-CoV-2 genome.
The two genomic sequences were closely related to SARS-CoV-2, with 92.6% identity across the genome. They also exhibited identical genomic organization. Thus, these two coronaviruses are a sublineage of SARS-CoV-2 related viruses, even though they are geographically distant.
A portion corresponding to the N terminal domain of the spike protein was dissimilar with SARS-CoV-2. This region was similar to more distantly related betacoronaviruses.
This data suggests that the ancestors of these viral sublineages co-circulate within a wider geographic area and more distinct bat species.
Receptor-binding domain
In silico structural analysis suggested that the external subdomain of the spike receptor-binding domain (RBD) structure is highly similar to SARS-CoV-2. Six amino acid residues determine efficient receptor binding of SARS-CoV-2 to the human angiotensin-converting enzyme 2 (hACE2) receptor. Five out of six amino acid residues are conserved.
The investigators then performed pseudovirus entry assays using HEK293T cells expressing either the bat ACE2 receptor or human ACE2 receptor. Viral pseudoparticles packaged a coronavirus spike from SARS-CoV-2 or the bat coronavirus. Pseudoviral particles expressing the bat coronavirus spike were not able to infect HEK293T cells expressing human ACE2 but they were able to infect HEK293T expressing bat ACE2. In addition, the pseudoviral particles expressing the SARS-CoV-2 spike were able to infect HEK293T cells expressing bat ACE2.
Conclusions
This study discovered SARS-CoV-2 related viruses in a bat species not found in China. This implies that these viruses have a much wider geographic distribution than previously reported. This study also suggests that Southeast Asia should be considered for future surveillance for coronaviruses.
Southeast Asia has a high diversity of wildlife. There also exists extensive trade in wild animals. Moreover, there is a dramatic land-use change due to infrastructure development, urban development, agricultural expansion, and human encroachment upon wildlife. Due to these factors, human contact with wild hosts of SARS-like coronaviruses has increased. Therefore, Southeast Asia may represent an area to consider for the ongoing research for the origins of SARS-CoV-2.
This study also suggests that Southeast Asia should be considered for future surveillance for coronaviruses.
According to this study, continued and expanded surveillance of bats and other wild animals in Southeast Asia is crucial for future pandemic preparedness and prevention.