Several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are highly infectious and can evade immunity induced by natural infection and vaccination.
 Study: Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike. Image Credit: Corona Borealis Studio / Shutterstock
Study: Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike. Image Credit: Corona Borealis Studio / Shutterstock
Introduction
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, several variants of concern (VOCs) have emerged. These variants are known to cause breakthrough infections, i.e., they can circumvent the protective effects of vaccination or infection-induced natural immunity. The neutralizing antibodies elicited by natural infection target the SARS-CoV-2 spike protein. Currently available COVID-19 vaccines also induce a neutralizing antibody response against the spike protein. The VOCs have mutations within the different regions of the spike protein.
Convalescent sera antibody levels and neutralization capabilities
The scientists collected blood samples from 10 convalescent donors around 49 days after symptom onset to analyze the specificity of individual memory B cells. Serum antibody reactivity was measured against spike proteins from the wildtype and D614G, B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.526 and B.1.617.1 variants and then compared.
Serum antibody levels from the 10 patients against wildtype and D614G spike antigens were similar. However, the antibody levels were reduced against the spike proteins of B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.526 and B.1.617.1 relative to the wildtype spike proteins.
Similarly, antibody levels against the receptor-binding domains (RBDs) of B.1.1.7, B.1.351 and P.1 were reduced relative to the wildtype RBD. However, there was a less than 2-fold decrease in antibody binding against single mutants of the RBD.
The sera retained similar neutralizing levels against the wildtype and the B.1.1.7 and P.1 SARS-CoV-2 variants. However, there was a reduction in neutralization against B.1.617.1 and B.1.617.2 compared to the wildtype.
These data indicate that serum antibodies elicited by natural infection were able to neutralize B.1.1.7, P.1 and wildtype viruses equally. Most donors lost neutralizing potential against B.1.617 lineage viruses.
Thus, convalescent-phase sera have reduced antibody titers but retain neutralization capabilities against circulating SARS-CoV-2 VOCs.
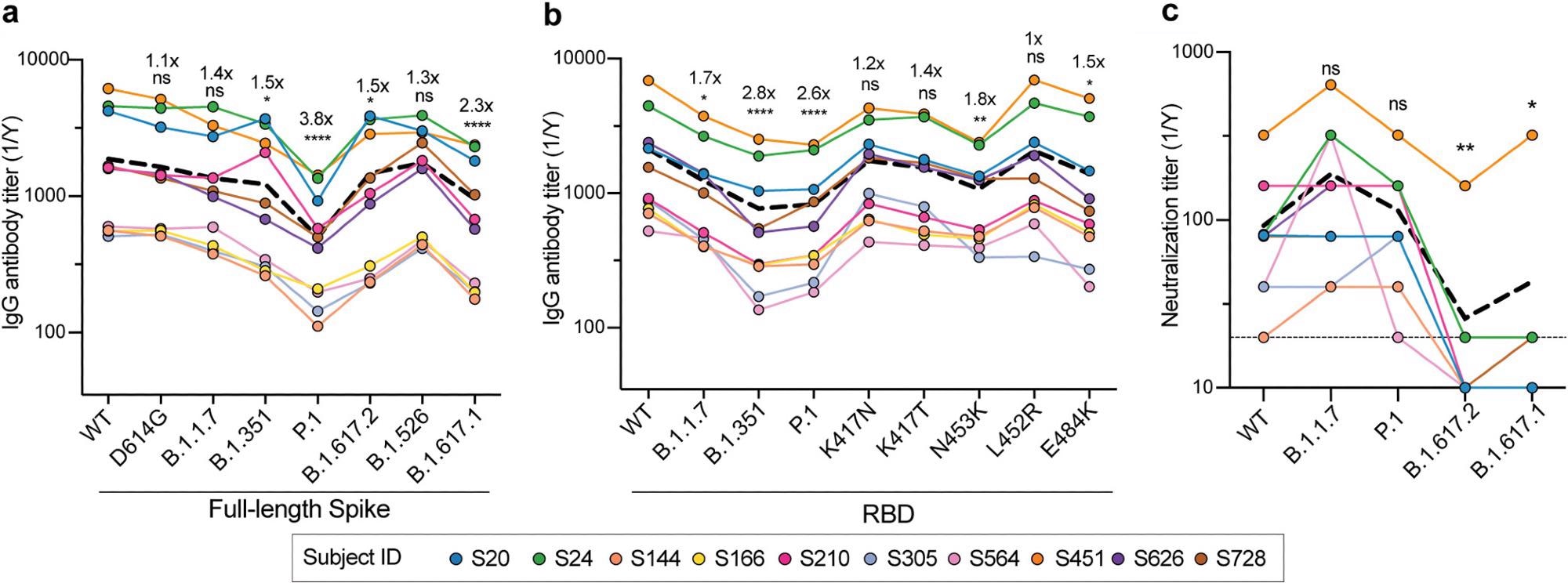 Analyses of serum antibody responses in COVID-19 convalescent individuals. (a and b) Total IgG endpoint antibody titers from 10 convalescent subjects against SARS-CoV-2 full-length spike variants (a) and RBD recombinant antigens (b). The dashed line is the mean IgG titer. (c) Neutralization titers from 10 convalescent donors against WT SARS-CoV-2, B.1.1.7, P.1, B.1.617.2, and B.1.617.1. The dashed line represents the mean neutralization titer. Data in panels a to c were analyzed using nonparametric Friedman’s test with Dunnett’s multiple-comparison test. Fold changes in relative mAb binding to variants or mutants compared to the WT in panels a and b are indicated above the statistical asterisks. ns, not significant.
Analyses of serum antibody responses in COVID-19 convalescent individuals. (a and b) Total IgG endpoint antibody titers from 10 convalescent subjects against SARS-CoV-2 full-length spike variants (a) and RBD recombinant antigens (b). The dashed line is the mean IgG titer. (c) Neutralization titers from 10 convalescent donors against WT SARS-CoV-2, B.1.1.7, P.1, B.1.617.2, and B.1.617.1. The dashed line represents the mean neutralization titer. Data in panels a to c were analyzed using nonparametric Friedman’s test with Dunnett’s multiple-comparison test. Fold changes in relative mAb binding to variants or mutants compared to the WT in panels a and b are indicated above the statistical asterisks. ns, not significant.
Monoclonal antibodies against distinct parts of the spike protein
The scientists then generated monoclonal antibodies (mAbs) from spike binding B cells isolated from 10 convalescent subjects. They sorted B cells binding to spike and/or RBD probes and performed single-cell RNA sequencing (RNA-seq) and B cell receptor sequencing.
The percentage of spike non-RBD binding B cells was 4-fold higher than that of RBD binding B cells. This means that natural infection preferentially induced B cell responses against regions on the spike outside the RBD.
The scientists generated 43 mAbs from all 10 donors specific for the WT spike protein. They investigated specific domain targeting by these mAbs using an enzyme-linked immunosorbent assay (ELISA). Eighteen out of 43 mAbs were neutralizing.
The mAbs binding the RBD and NTD-B were neutralizing and mAbs binding NTD-A and S2 were non-neutralizing. Eight mAbs were potently neutralizing antibodies and three out of seven NTD-B mAbs had moderate neutralization potency.
Thus, mAbs against the RBD were the predominant source of neutralizing antibodies induced by wildtype SARS-CoV-2 infection.
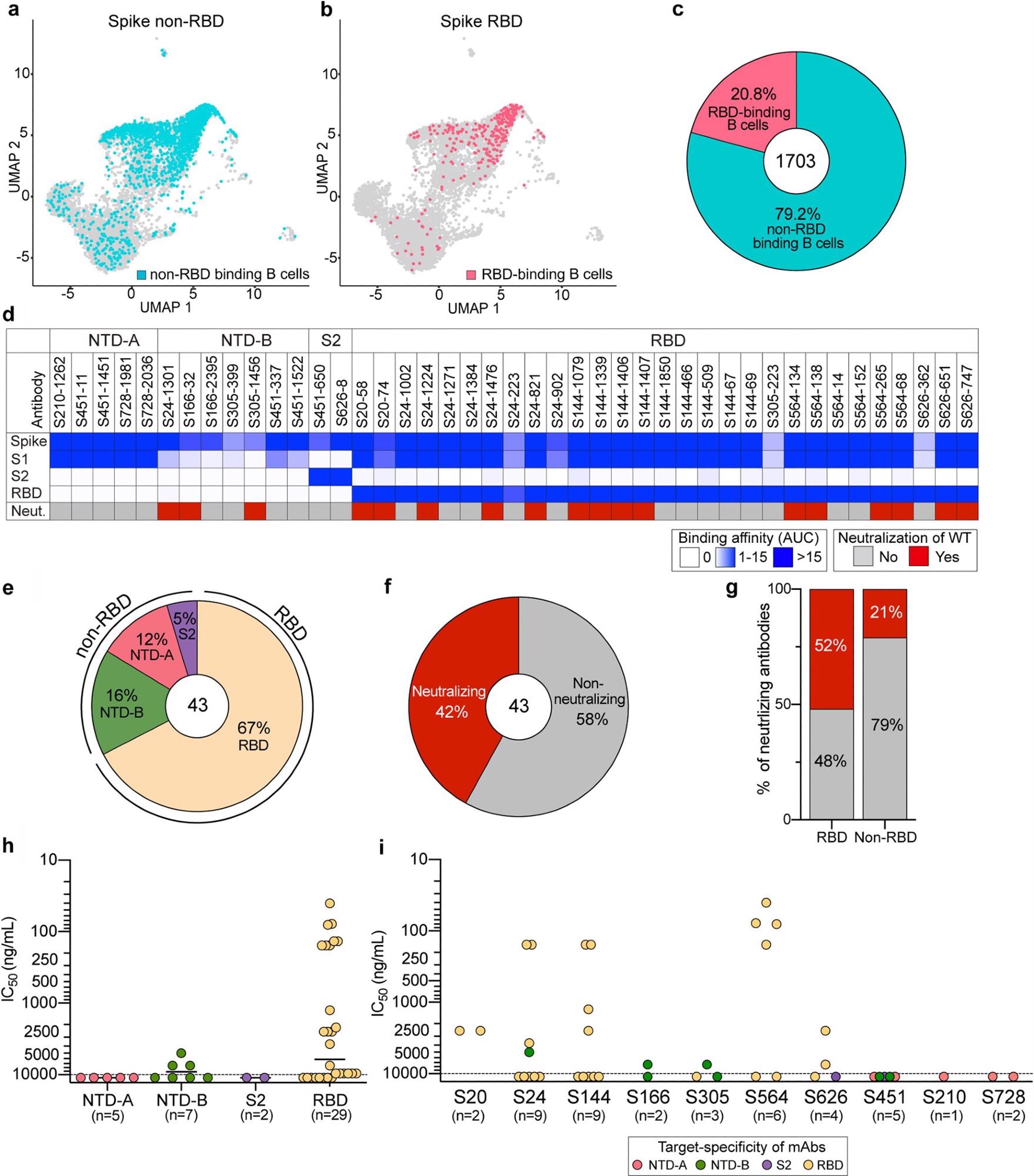 Characterization of spike-reactive mAbs. (a and b) Uniform manifold approximation and projection (UMAP) of SARS-CoV-2 spike non-RBD binding (a) and spike RBD binding (b) B cells isolated from PBMCs of 10 convalescent subjects. (c) Proportion of spike non-RBD- and spike RBD-specific binding B cells. The number in the center of the pie chart indicates the number of antigen-specific binding B cells. (d) mAbs generated from selected B cells (n = 43) were tested for binding to full-length spike, S1, S2, and the RBD and neutralization potential against WT SARS-CoV-2. Binding data are represented as areas under the curve (AUC). Neutralizing activities of <10,000 ng/ml are considered neutralizing. (e and f) Pie charts of mAb domain specificity (e) and neutralizing capability (f). The numbers in the center of the pie graphs indicate the number of antibodies tested. (g) Comparison of neutralizing capabilities of mAbs targeting the spike RBD and spike non-RBD. (h and i) IC50s of the neutralization potencies of spike-reactive antibodies against WT virus based on domain specificity (h) and by subject (i). The mean in panel h is indicated as a solid line. Data in panels h and i are colored based on domain specificity, and the dashed lines shown in panels h and i indicate the limit of detection (10,000 ng/ml). Data in panels d to i are representative of results from two independent experiments performed in duplicate.
Characterization of spike-reactive mAbs. (a and b) Uniform manifold approximation and projection (UMAP) of SARS-CoV-2 spike non-RBD binding (a) and spike RBD binding (b) B cells isolated from PBMCs of 10 convalescent subjects. (c) Proportion of spike non-RBD- and spike RBD-specific binding B cells. The number in the center of the pie chart indicates the number of antigen-specific binding B cells. (d) mAbs generated from selected B cells (n = 43) were tested for binding to full-length spike, S1, S2, and the RBD and neutralization potential against WT SARS-CoV-2. Binding data are represented as areas under the curve (AUC). Neutralizing activities of <10,000 ng/ml are considered neutralizing. (e and f) Pie charts of mAb domain specificity (e) and neutralizing capability (f). The numbers in the center of the pie graphs indicate the number of antibodies tested. (g) Comparison of neutralizing capabilities of mAbs targeting the spike RBD and spike non-RBD. (h and i) IC50s of the neutralization potencies of spike-reactive antibodies against WT virus based on domain specificity (h) and by subject (i). The mean in panel h is indicated as a solid line. Data in panels h and i are colored based on domain specificity, and the dashed lines shown in panels h and i indicate the limit of detection (10,000 ng/ml). Data in panels d to i are representative of results from two independent experiments performed in duplicate.
Cross-reactive mAbs
The scientists tested non-RBD-targeting mAbs for binding to a panel of SARS-CoV-2 variants. All non-RBD spike-reactive antibodies showed similar binding to the D614G spike.
All mAbs targeting NTD-A and S2 maintained similar binding to the spike of the B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.526, and B.1.617.1 variants.
The NTD-B mAbs showed significantly reduced binding to the spike of B.1.1.7, B.1.351, B.1.617.2, and B.1.617.1 while showing similar binding to B.1.526 and a minor reduction in binding to the spike of P.1.
The antibodies against NTD-B showed cross-neutralization capacity and thus may provide protection against some emerging VOCs such as B.1.1.7 and P.1.
The scientists tested RBD-targeting mAbs for binding to RBD mutants that had a single mutation. They also tested mAb binding to the RBDs of SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) to investigate cross-reactivity to other coronaviruses.
Six mAbs could not be classified because they either lost binding to multiple mutant classes or bound equally to all RBD mutants but did not bind to SARS-CoV-1 or MERS-CoV.
A subset of RBD-binding mAbs retained neutralization activity against VOCs. None of the neutralizing mAbs induced by natural wildtype infection neutralized all emerging SARS-CoV-2 variants. However, one mAb neutralized each VOC, suggesting that the convalescent donors generated a diverse cross-neutralizing antibody response. Therefore, antibodies targeting multiple regions on the spike protein are important against emerging VOCs.
The majority of antibodies isolated from donors with high antibody levels had lower neutralizing potency than antibodies derived from donors who had lower antibody levels and less severity.
Conclusion
SARS-CoV-2 infection induces cross-neutralizing immunity against circulating VOCs. Most probably, this is due to antibodies targeting many different regions of the spike protein.
This study suggests that different cross-neutralizing antibodies that bind specific sites on the spike protein should be induced to protect against SARS-CoV-2 variants and limit the virus from escaping any single antibody target.
Journal reference:
- Siriruk C, Yanbin F, J. GJ, et al. (2021) Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike. mBio, 0(0), e02975-21. https://doi.org/10.1128/mBio.02975-21